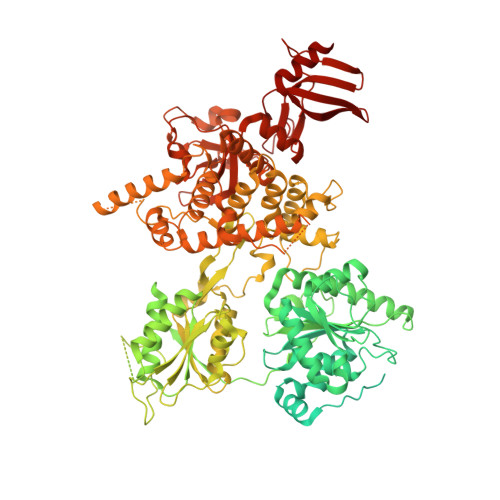
The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis.
Roychowdhury, A., Joret, C., Bourgeois, G., Heurgue-Hamard, V., Lafontaine, D.L.J., Graille, M.(2019) Nucleic Acids Res 47: 7548-7563
- PubMed: 31188444
- DOI: https://doi.org/10.1093/nar/gkz529
- Primary Citation of Related Structures:
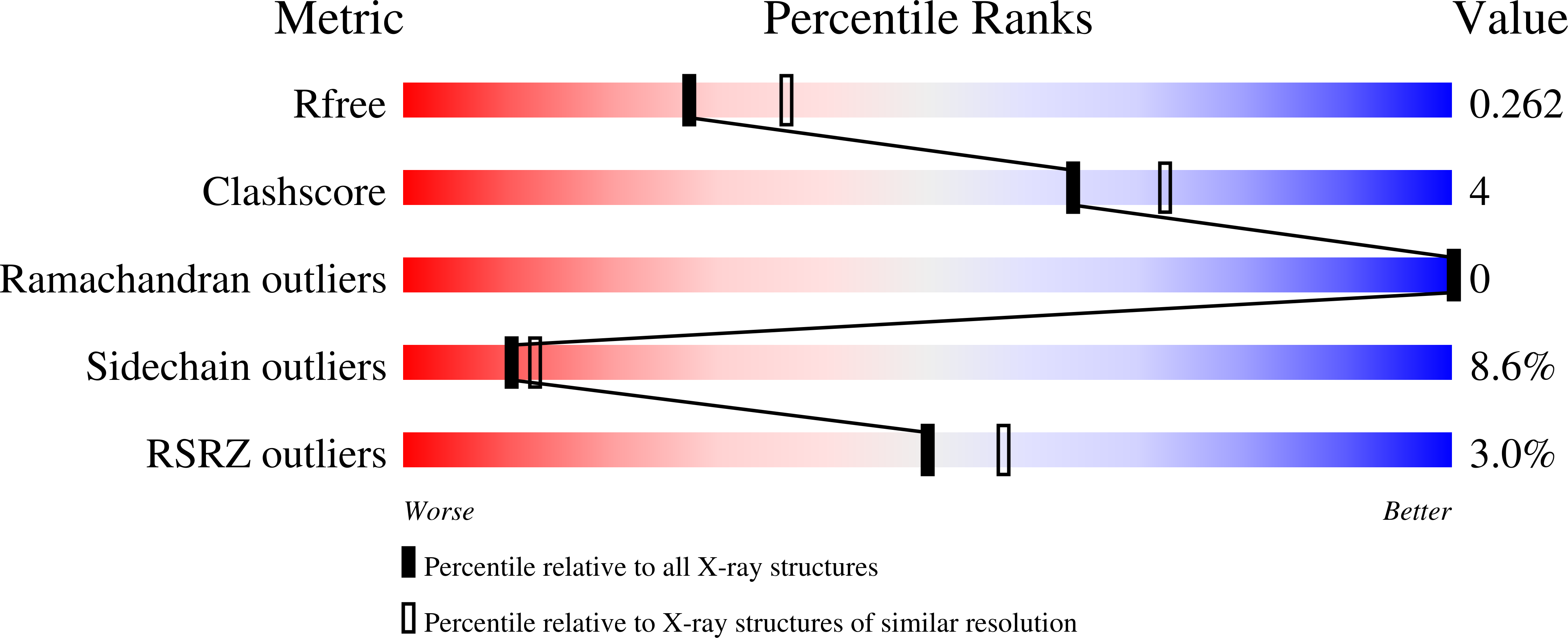
6H57 - PubMed Abstract:
Ribosome biogenesis is an essential process in all living cells, which entails countless highly sequential and dynamic structural reorganization events. These include formation of dozens RNA helices through Watson-Crick base-pairing within ribosomal RNAs (rRNAs) and between rRNAs and small nucleolar RNAs (snoRNAs), transient association of hundreds of proteinaceous assembly factors to nascent precursor (pre-)ribosomes, and stable assembly of ribosomal proteins. Unsurprisingly, the largest group of ribosome assembly factors are energy-consuming proteins (NTPases) including 25 RNA helicases in budding yeast. Among these, the DEAH-box Dhr1 is essential to displace the box C/D snoRNA U3 from the pre-rRNAs where it is bound in order to prevent premature formation of the central pseudoknot, a dramatic irreversible long-range interaction essential to the overall folding of the small ribosomal subunit. Here, we report the crystal structure of the Dhr1 helicase module, revealing the presence of a remarkable carboxyl-terminal domain essential for Dhr1 function in ribosome biogenesis in vivo and important for its interaction with its coactivator Utp14 in vitro. Furthermore, we report the functional consequences on ribosome biogenesis of DHX37 (human Dhr1) mutations found in patients suffering from microcephaly and other neurological diseases.
Organizational Affiliation:
BIOC, CNRS, Ecole polytechnique, IP Paris, F-91128 Palaiseau, France.