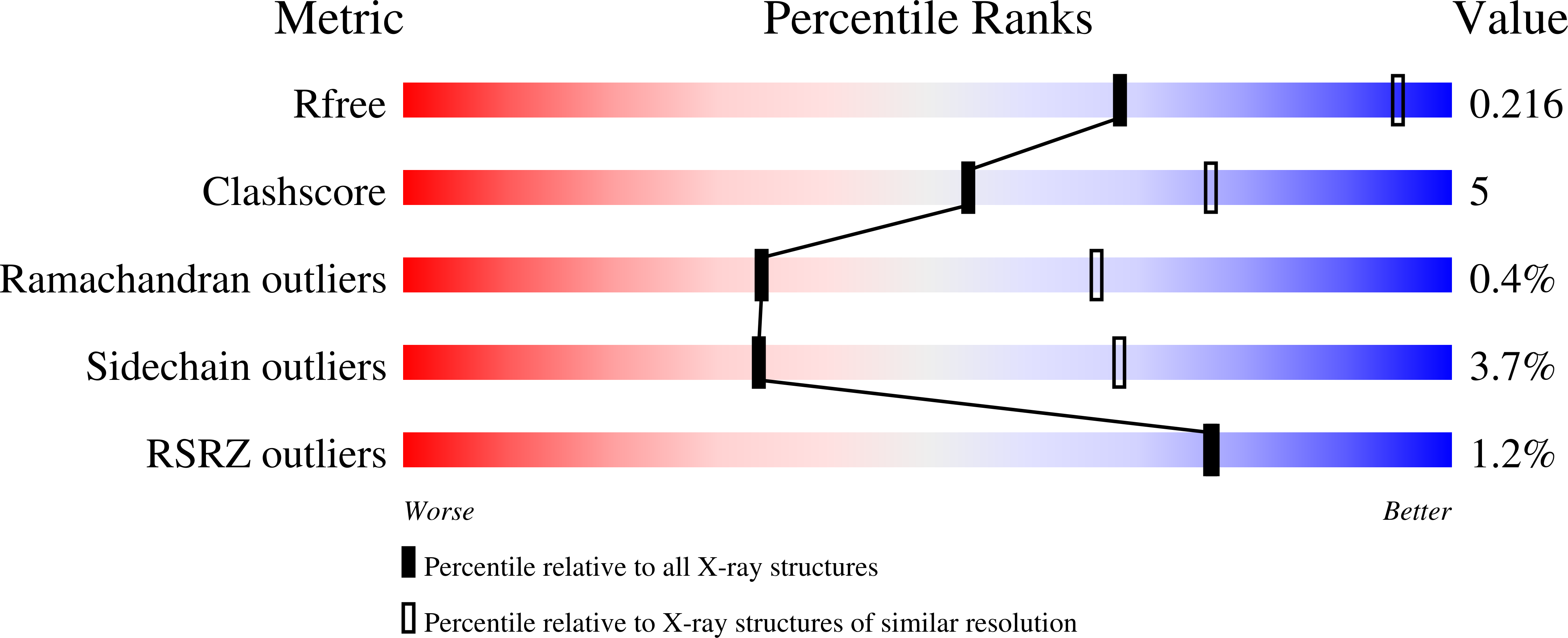
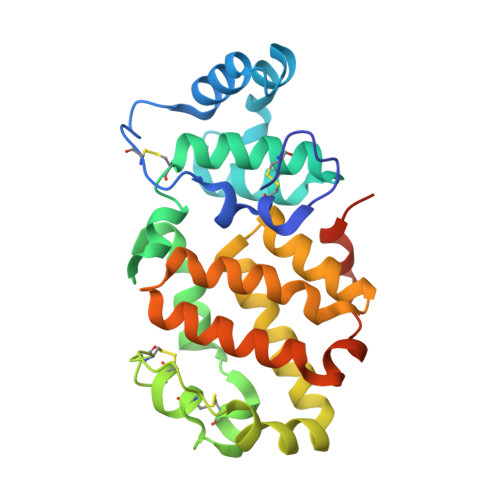
Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Hellert, J., Aebischer, A., Wernike, K., Haouz, A., Brocchi, E., Reiche, S., Guardado-Calvo, P., Beer, M., Rey, F.A.(2019) Nat Commun 10: 879-879
- PubMed: 30787296
- DOI: https://doi.org/10.1038/s41467-019-08832-8
- Primary Citation of Related Structures:
6H3S, 6H3T, 6H3U, 6H3V, 6H3W, 6H3X - PubMed Abstract:
Orthobunyaviruses (OBVs) form a distinct genus of arthropod-borne bunyaviruses that can cause severe disease upon zoonotic transmission to humans. Antigenic drift or genome segment re-assortment have in the past resulted in new pathogenic OBVs, making them potential candidates for causing emerging zoonoses in the future. Low-resolution electron cryo-tomography studies have shown that OBV particles feature prominent trimeric spikes, but their molecular organization remained unknown. Here we report X-ray crystallography studies of four different OBVs showing that the spikes are formed by an N-terminal extension of the fusion glycoprotein Gc. Using Schmallenberg virus, a recently emerged OBV, we also show that the projecting spike is the major target of the neutralizing antibody response, and provide X-ray structures in complex with two protecting antibodies. We further show that immunization of mice with the spike domains elicits virtually sterilizing immunity, providing fundamental knowledge essential in the preparation for potential newly emerging OBV zoonoses.
Organizational Affiliation:
Structural Virology Unit, Virology Department, Institut Pasteur, CNRS UMR 3569, 25-28 rue du Dr. Roux, 75015, Paris, France.