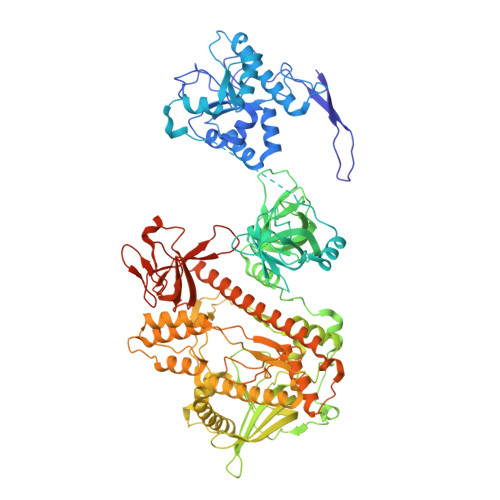
Distinct and evolutionary conserved structural features of the human nuclear exosome complex.
Gerlach, P., Schuller, J.M., Bonneau, F., Basquin, J., Reichelt, P., Falk, S., Conti, E.(2018) Elife 7
- PubMed: 30047866
- DOI: https://doi.org/10.7554/eLife.38686
- Primary Citation of Related Structures:
6H25 - PubMed Abstract:
The nuclear RNA exosome complex mediates the processing of structured RNAs and the decay of aberrant non-coding RNAs, an important function particularly in human cells. Most mechanistic studies to date have focused on the yeast system. Here, we reconstituted and studied the properties of a recombinant 14-subunit human nuclear exosome complex. In biochemical assays, the human exosome embeds a longer RNA channel than its yeast counterpart. The 3.8 Å resolution cryo-EM structure of the core complex bound to a single-stranded RNA reveals that the RNA channel path is formed by two distinct features of the hDIS3 exoribonuclease: an open conformation and a domain organization more similar to bacterial RNase II than to yeast Rrp44. The cryo-EM structure of the holo-complex shows how obligate nuclear cofactors position the hMTR4 helicase at the entrance of the core complex, suggesting a striking structural conservation from lower to higher eukaryotes.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Munich, Germany.