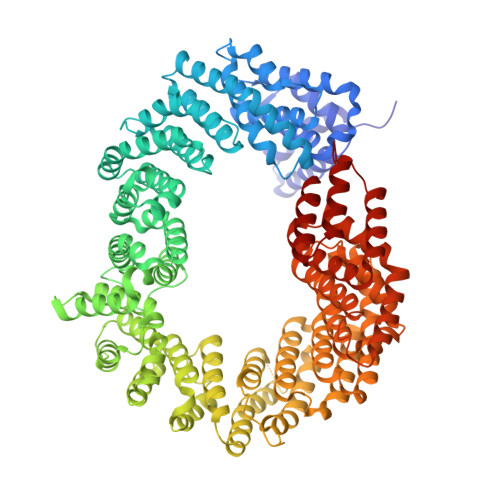
Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Jang, S., Cook, N.J., Pye, V.E., Bedwell, G.J., Dudek, A.M., Singh, P.K., Cherepanov, P., Engelman, A.N.(2019) Nucleic Acids Res 47: 4663-4683
- PubMed: 30916345
- DOI: https://doi.org/10.1093/nar/gkz206
- Primary Citation of Related Structures:
6GX9 - PubMed Abstract:
Cleavage factor I mammalian (CFIm) complex, composed of cleavage and polyadenylation specificity factor 5 (CPSF5) and serine/arginine-like protein CPSF6, regulates alternative polyadenylation (APA). Loss of CFIm function results in proximal polyadenylation site usage, shortening mRNA 3' untranslated regions (UTRs). Although CPSF6 plays additional roles in human disease, its nuclear translocation mechanism remains unresolved. Two β-karyopherins, transportin (TNPO) 1 and TNPO3, can bind CPSF6 in vitro, and we demonstrate here that while the TNPO1 binding site is dispensable for CPSF6 nuclear import, the arginine/serine (RS)-like domain (RSLD) that mediates TNPO3 binding is critical. The crystal structure of the RSLD-TNPO3 complex revealed potential CPSF6 interaction residues, which were confirmed to mediate TNPO3 binding and CPSF6 nuclear import. Both binding and nuclear import were independent of RSLD phosphorylation, though a hyperphosphorylated mimetic mutant failed to bind TNPO3 and mislocalized to the cell cytoplasm. Although hypophosphorylated CPSF6 largely supported normal polyadenylation site usage, a significant number of mRNAs harbored unnaturally extended 3' UTRs, similar to what is observed when other APA regulators, such as CFIIm component proteins, are depleted. Our results clarify the mechanism of CPSF6 nuclear import and highlight differential roles for RSLD phosphorylation in nuclear translocation versus regulation of APA.
Organizational Affiliation:
Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute, Boston, MA 02215, USA.