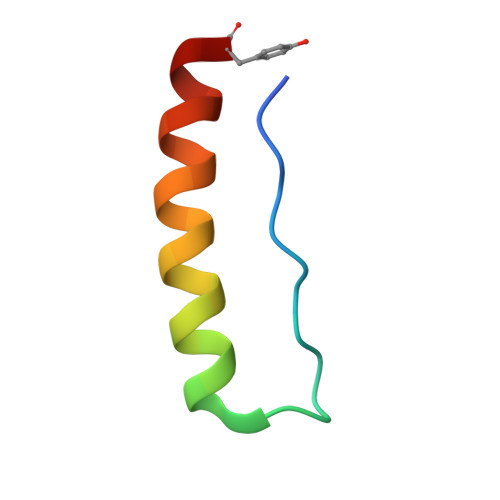
Stabilizing and Understanding a Miniprotein by Rational Redesign.
Porter Goff, K.L., Nicol, D., Williams, C., Crump, M.P., Zieleniewski, F., Samphire, J.L., Baker, E.G., Woolfson, D.N.(2019) Biochemistry 58: 3060-3064
- PubMed: 31251570
- DOI: https://doi.org/10.1021/acs.biochem.9b00067
- Primary Citation of Related Structures:
6GWX - PubMed Abstract:
Miniproteins reduce the complexity of the protein-folding problem allowing systematic studies of contributions to protein folding and stabilization. Here, we describe the rational redesign of a miniprotein, PPα, comprising a polyproline II helix, a loop, and an α helix. The redesign provides a de novo framework for interrogating noncovalent interactions. Optimized PPα has significantly improved thermal stability with a midpoint unfolding temperature ( T M ) of 51 °C. Its nuclear magnetic resonance structure indicates a density of stabilizing noncovalent interactions that is higher than that of the parent peptide, specifically an increased number of CH-π interactions. In part, we attribute this to improved long-range electrostatic interactions between the two helical elements. We probe further sequence-stability relationships in the miniprotein through a series of rational mutations.
Organizational Affiliation:
School of Chemistry , University of Bristol , Cantock's Close , Bristol BS8 1TS , U.K.