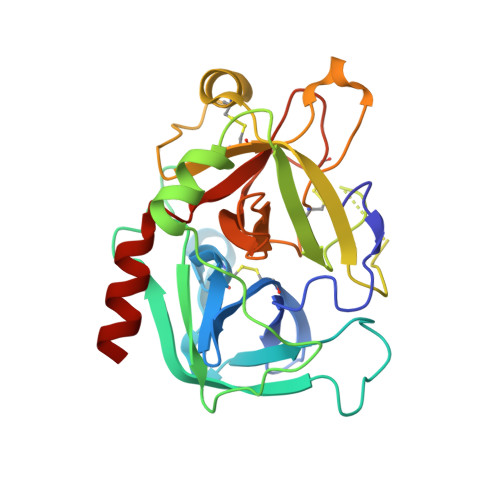
Thiol-to-amine cyclization reaction enables screening of large libraries of macrocyclic compounds and the generation of sub-kilodalton ligands.
Kale, S.S., Bergeron-Brlek, M., Wu, Y., Kumar, M.G., Pham, M.V., Bortoli, J., Vesin, J., Kong, X.D., Machado, J.F., Deyle, K., Gonschorek, P., Turcatti, G., Cendron, L., Angelini, A., Heinis, C.(2019) Sci Adv 5: eaaw2851-eaaw2851
- PubMed: 31457083
- DOI: https://doi.org/10.1126/sciadv.aaw2851
- Primary Citation of Related Structures:
6GWE - PubMed Abstract:
Macrocyclic compounds are an attractive modality for drug development, but the limited availability of large, structurally diverse macrocyclic libraries hampers the discovery of leads. Here, we describe the discovery of efficient macrocyclization reactions based on thiol-to-amine ligations using bis-electrophiles, their application to synthesize and screen large libraries of macrocyclic compounds, and the identification of potent small macrocyclic ligands. The thiol-to-amine cyclization reactions showed unexpectedly high yields for a wide substrate range, which obviated product purification and enabled the generation and screening of an 8988 macrocycle library with a comparatively small effort. X-ray structure analysis of an identified thrombin inhibitor ( K i = 42 ± 5 nM) revealed a snug fit with the target, validating the strategy of screening large libraries with a high skeletal diversity. The approach provides a route for screening large sub-kilodalton macrocyclic libraries and may be applied to many challenging drug targets.
Organizational Affiliation:
Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.