Structural and biochemical characterization of the Cutibacterium acnes exo-beta-1,4-mannosidase that targets the N-glycan core of host glycoproteins.
Reichenbach, T., Kalyani, D., Gandini, R., Svartstrom, O., Aspeborg, H., Divne, C.(2018) PLoS One 13: e0204703-e0204703
- PubMed: 30261037
- DOI: https://doi.org/10.1371/journal.pone.0204703
- Primary Citation of Related Structures:
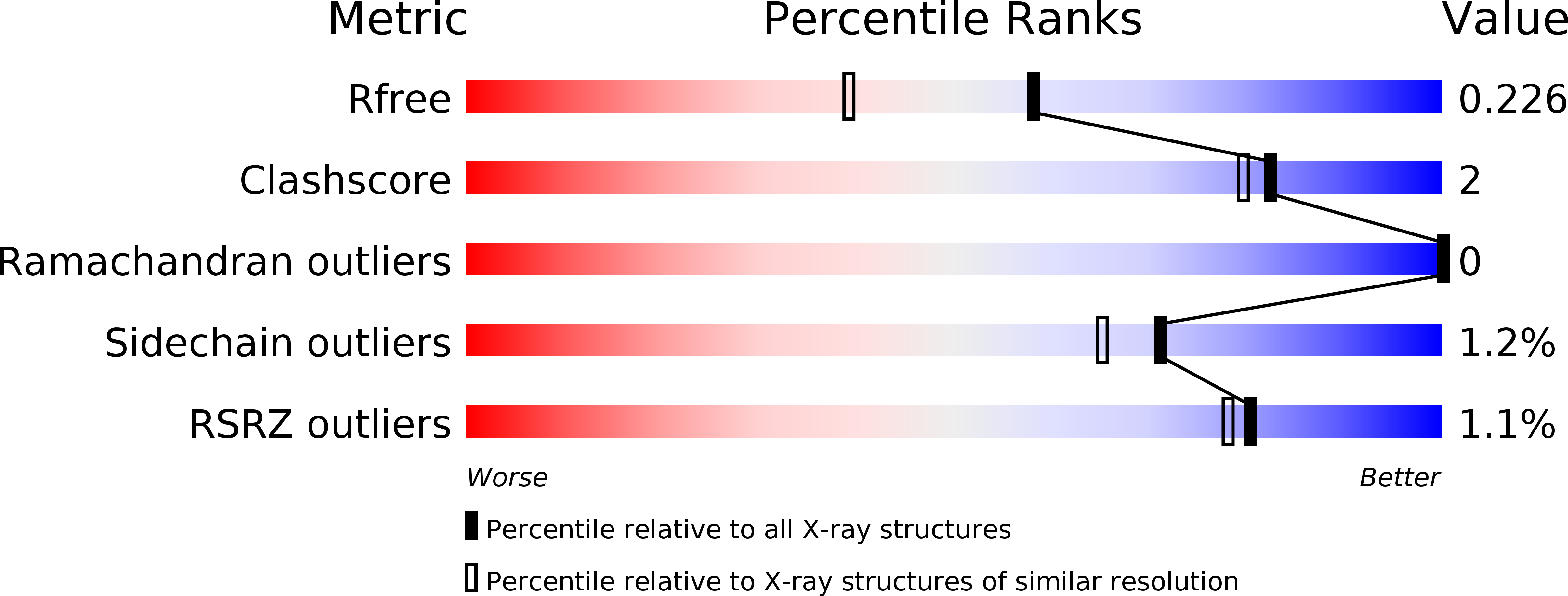
6GVB - PubMed Abstract:
Commensal and pathogenic bacteria have evolved efficient enzymatic pathways to feed on host carbohydrates, including protein-linked glycans. Most proteins of the human innate and adaptive immune system are glycoproteins where the glycan is critical for structural and functional integrity. Besides enabling nutrition, the degradation of host N-glycans serves as a means for bacteria to modulate the host's immune system by for instance removing N-glycans on immunoglobulin G. The commensal bacterium Cutibacterium acnes is a gram-positive natural bacterial species of the human skin microbiota. Under certain circumstances, C. acnes can cause pathogenic conditions, acne vulgaris, which typically affects 80% of adolescents, and can become critical for immunosuppressed transplant patients. Others have shown that C. acnes can degrade certain host O-glycans, however, no degradation pathway for host N-glycans has been proposed. To investigate this, we scanned the C. acnes genome and were able to identify a set of gene candidates consistent with a cytoplasmic N-glycan-degradation pathway of the canonical eukaryotic N-glycan core. We also found additional gene sequences containing secretion signals that are possible candidates for initial trimming on the extracellular side. Furthermore, one of the identified gene products of the cytoplasmic pathway, AEE72695, was produced and characterized, and found to be a functional, dimeric exo-β-1,4-mannosidase with activity on the β-1,4 glycosidic bond between the second N-acetylglucosamine and the first mannose residue in the canonical eukaryotic N-glycan core. These findings corroborate our model of the cytoplasmic part of a C. acnes N-glycan degradation pathway.
Organizational Affiliation:
Department of Industrial Biotechnology, School of Engineering Sciences in Chemistry, Biotechnology, and Health (CBH), KTH Royal Institute of Technology, Stockholm, Sweden.