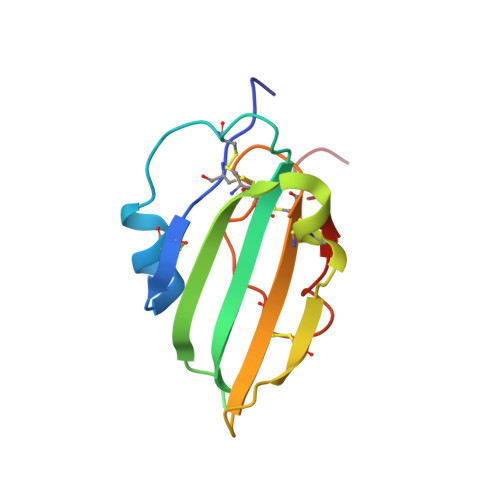
Structure of the Wnt signaling enhancer LYPD6 and its interactions with the Wnt coreceptor LRP6.
Zhao, Y., Ren, J., Lu, W., Harlos, K., Jones, E.Y.(2018) FEBS Lett 592: 3152-3162
- PubMed: 30069874
- DOI: https://doi.org/10.1002/1873-3468.13212
- Primary Citation of Related Structures:
6GBI - PubMed Abstract:
Ly6/urokinase-type plasminogen activator receptor (uPAR) (LU) domain containing 6 (LYPD6) is a Wnt signaling enhancer that promotes phosphorylation of the Wnt coreceptor low density lipoprotein receptor-related protein 6 (LRP6). It also binds the nicotinic acetylcholine receptor (nAChR). We report here the 1.25 Å resolution structure of the LYPD6 extracellular LU domain and map its interaction with LRP6 by mutagenesis and surface plasmon resonance. The LYPD6 LU structure reveals a 'trifingered protein domain' fold with the middle fingertip bearing an 'NxI' motif, a tripeptide motif associated with LRP5/6 binding by Wnt inhibitors. Of the Ly6 protein family members, only LYPD6 has an NxI motif. Since mutations in the LYPD6 NxI motif abolish or severely reduce interaction with LRP6, our results indicate its key role in the interaction of LYPD6 with LRP6.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, UK.