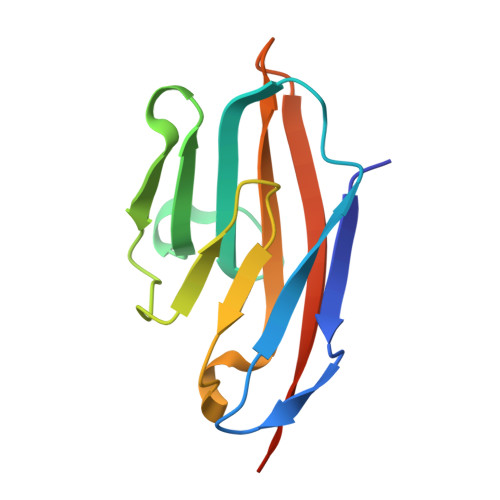
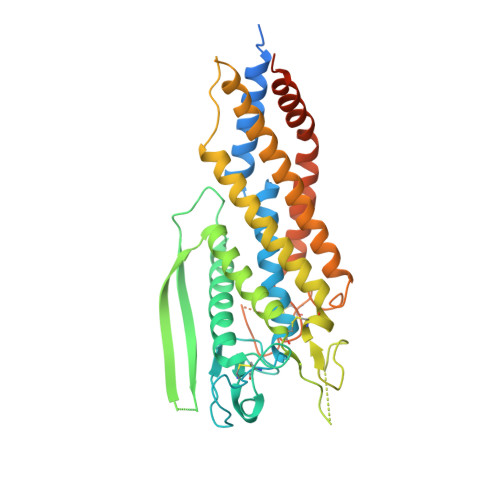
Helicobacter pyloriadhesin HopQ disruptstransdimerization in human CEACAMs.
Moonens, K., Hamway, Y., Neddermann, M., Reschke, M., Tegtmeyer, N., Kruse, T., Kammerer, R., Mejias-Luque, R., Singer, B.B., Backert, S., Gerhard, M., Remaut, H.(2018) EMBO J 37
- PubMed: 29858229
- DOI: https://doi.org/10.15252/embj.201798665
- Primary Citation of Related Structures:
6GBG, 6GBH - PubMed Abstract:
The human gastric pathogen Helicobacter pylori is a major causative agent of gastritis, peptic ulcer disease, and gastric cancer. As part of its adhesive lifestyle, the bacterium targets members of the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family by the conserved outer membrane adhesin HopQ. The HopQ-CEACAM1 interaction is associated with inflammatory responses and enables the intracellular delivery and phosphorylation of the CagA oncoprotein via a yet unknown mechanism. Here, we generated crystal structures of HopQ isotypes I and II bound to the N-terminal domain of human CEACAM1 (C1ND) and elucidated the structural basis of H. pylori specificity toward human CEACAM receptors. Both HopQ alleles target the β-strands G, F, and C of C1ND, which form the trans dimerization interface in homo- and heterophilic CEACAM interactions. Using SAXS, we show that the HopQ ectodomain is sufficient to induce C1ND monomerization and thus providing H. pylori a route to influence CEACAM-mediated cell adherence and signaling events.
Organizational Affiliation:
Structural and Molecular Microbiology, Structural Biology Research Center, VIB, Brussels, Belgium.