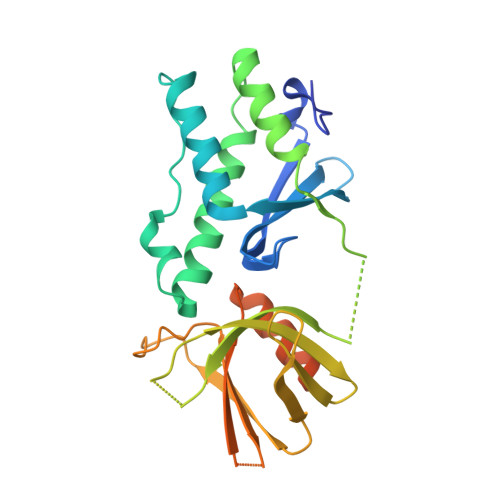
Structure of the tandem PX-PH domains of Bem3 from Saccharomyces cerevisiae.
Ali, I., Eu, S., Koch, D., Bleimling, N., Goody, R.S., Muller, M.P.(2018) Acta Crystallogr F Struct Biol Commun 74: 315-321
- PubMed: 29718000
- DOI: https://doi.org/10.1107/S2053230X18005915
- Primary Citation of Related Structures:
6FSF - PubMed Abstract:
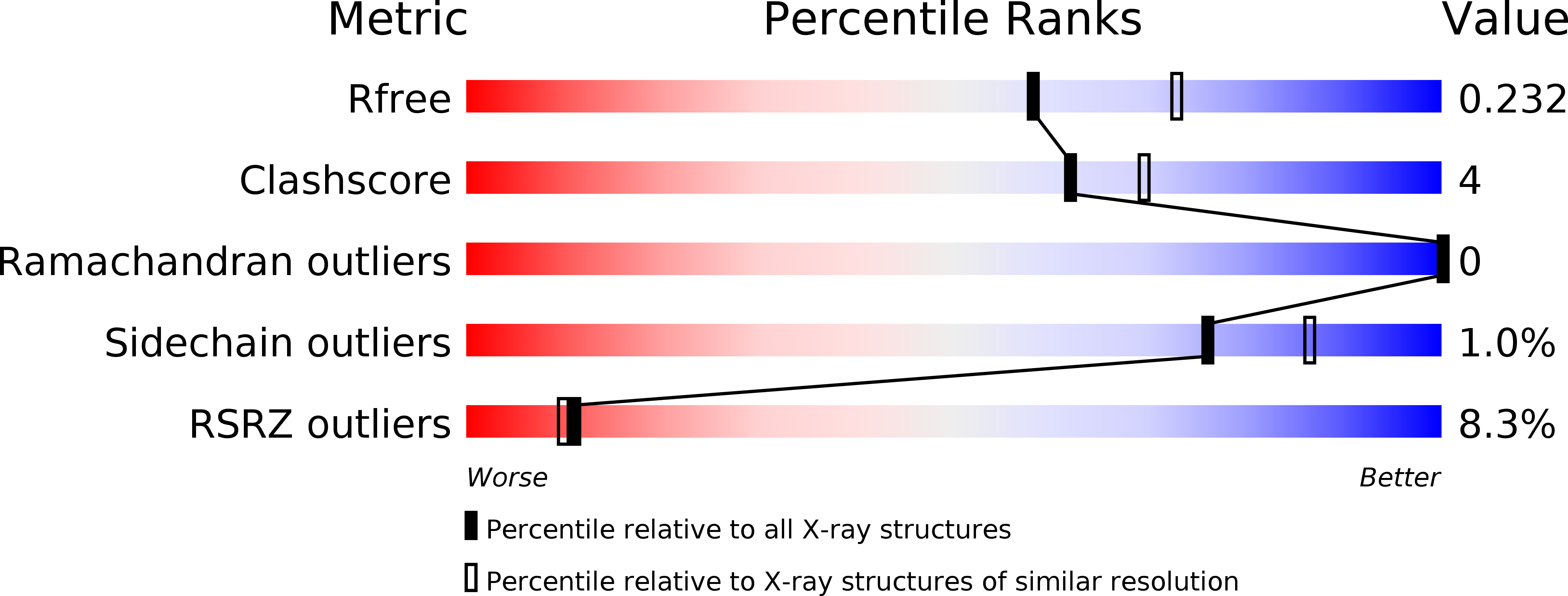
The structure of the tandem lipid-binding PX and pleckstrin-homology (PH) domains of the Cdc42 GTPase-activating protein Bem3 from Saccharomyces cerevisiae (strain S288c) has been determined to a resolution of 2.2 Å (R work = 21.1%, R free = 23.4%). It shows that the domains adopt a relative orientation that enables them to simultaneously bind to a membrane and suggests possible cooperativity in membrane binding.
Organizational Affiliation:
Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.