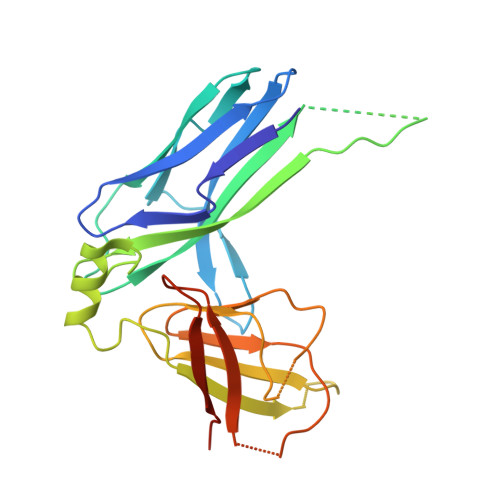
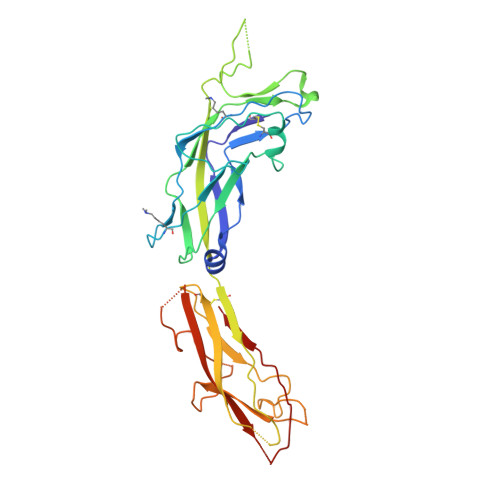
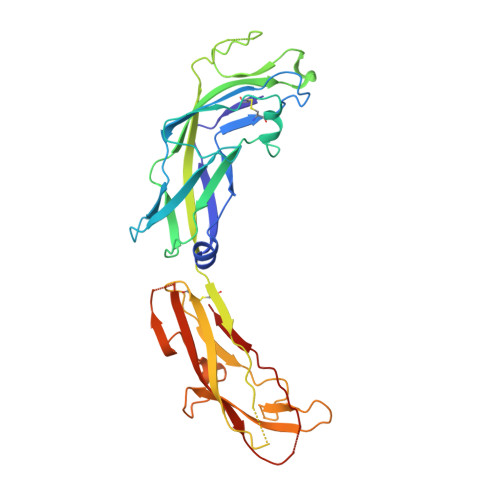
Structural basis forAcinetobacter baumanniibiofilm formation.
Pakharukova, N., Tuittila, M., Paavilainen, S., Malmi, H., Parilova, O., Teneberg, S., Knight, S.D., Zavialov, A.V.(2018) Proc Natl Acad Sci U S A 115: 5558-5563
- PubMed: 29735695
- DOI: https://doi.org/10.1073/pnas.1800961115
- Primary Citation of Related Structures:
6FJY - PubMed Abstract:
Acinetobacter baumannii -a leading cause of nosocomial infections-has a remarkable capacity to persist in hospital environments and medical devices due to its ability to form biofilms. Biofilm formation is mediated by Csu pili, assembled via the "archaic" chaperone-usher pathway. The X-ray structure of the CsuC-CsuE chaperone-adhesin preassembly complex reveals the basis for bacterial attachment to abiotic surfaces. CsuE exposes three hydrophobic finger-like loops at the tip of the pilus. Decreasing the hydrophobicity of these abolishes bacterial attachment, suggesting that archaic pili use tip-fingers to detect and bind to hydrophobic cavities in substrates. Antitip antibody completely blocks biofilm formation, presenting a means to prevent the spread of the pathogen. The use of hydrophilic materials instead of hydrophobic plastics in medical devices may represent another simple and cheap solution to reduce pathogen spread. Phylogenetic analysis suggests that the tip-fingers binding mechanism is shared by all archaic pili carrying two-domain adhesins. The use of flexible fingers instead of classical receptor-binding cavities is presumably more advantageous for attachment to structurally variable substrates, such as abiotic surfaces.
Organizational Affiliation:
Department of Chemistry, University of Turku, Joint Biotechnology Laboratory, Arcanum, 20500 Turku, Finland.