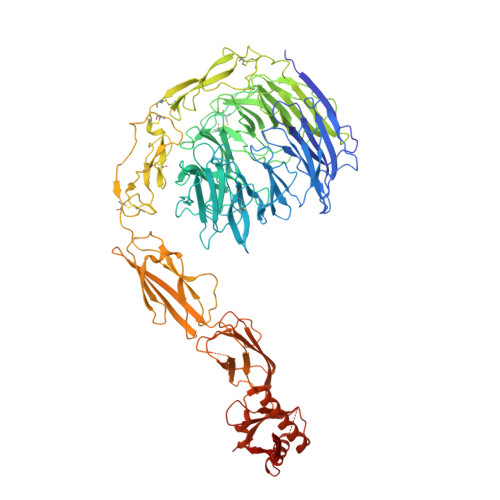
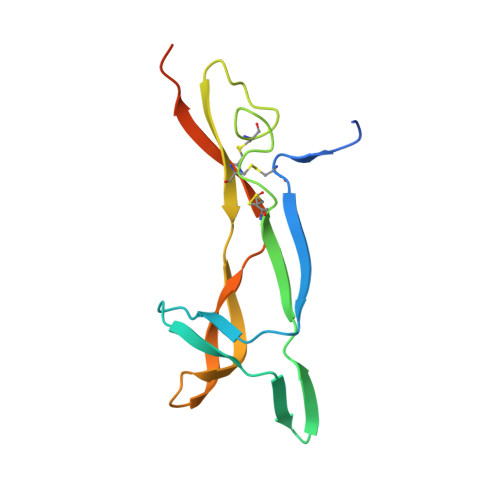
Structural insights into SorCS2-Nerve Growth Factor complex formation.
Leloup, N., Chataigner, L.M.P., Janssen, B.J.C.(2018) Nat Commun 9: 2979-2979
- PubMed: 30061605
- DOI: https://doi.org/10.1038/s41467-018-05405-z
- Primary Citation of Related Structures:
6FFY, 6FG9 - PubMed Abstract:
Signaling of SorCS receptors by proneurotrophin ligands regulates neuronal plasticity, induces apoptosis and is associated with mental disorders. The detailed structure of SorCS2 and its extracellular specificity are unresolved. Here we report crystal structures of the SorCS2-NGF complex and unliganded SorCS2 ectodomain, revealing cross-braced SorCS2 homodimers with two NGF dimers bound in a 2:4 stoichiometry. Five out of six SorCS2 domains directly contribute to dimer formation and a C-terminal membrane proximal unreported domain, with an RNA recognition motif fold, locks the dimer in an intermolecular head-to-tail interaction. The complex structure shows an altered SorCS2 conformation indicating substantial structural plasticity. Both NGF dimer chains interact exclusively with the top face of a SorCS2 β-propeller. Biophysical experiments reveal that NGF, proNGF, and proBDNF bind at this site on SorCS2. Taken together, our data reveal a structurally flexible SorCS2 receptor that employs the large β-propeller as a ligand binding platform.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Faculty of Science, Utrecht University, 3584 CH, Utrecht, The Netherlands.