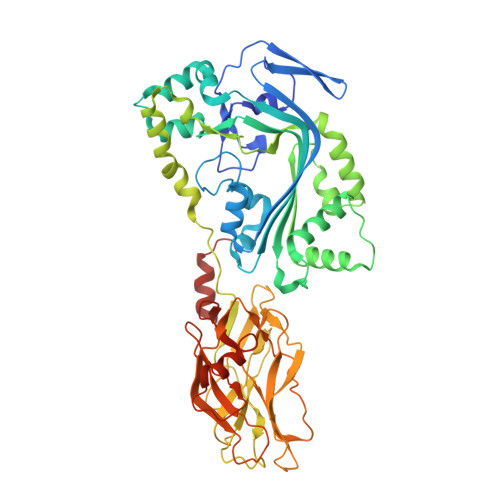
Structure-function characterization of an insecticidal protein GNIP1Aa, a member of an MACPF and beta-tripod families.
Zaitseva, J., Vaknin, D., Krebs, C., Doroghazi, J., Milam, S.L., Balasubramanian, D., Duck, N.B., Freigang, J.(2019) Proc Natl Acad Sci U S A 116: 2897-2906
- PubMed: 30728296
- DOI: https://doi.org/10.1073/pnas.1815547116
- Primary Citation of Related Structures:
6FBM - PubMed Abstract:
The crystal structure of the Gram-negative insecticidal protein, GNIP1Aa, has been solved at 2.5-Å resolution. The protein consists of two structurally distinct domains, a MACPF (membrane attack complex/PerForin) and a previously uncharacterized type of domain. GNIP1Aa is unique in being a prokaryotic MACPF member to have both its structure and function identified. It was isolated from a Chromobacterium piscinae strain and is specifically toxic to Diabrotica virgifera virgifera larvae upon feeding. In members of the MACPF family, the MACPF domain has been shown to be important for protein oligomerization and formation of transmembrane pores, while accompanying domains define the specificity of the target of the toxicity. In GNIP1Aa the accompanying C-terminal domain has a unique fold composed of three pseudosymmetric subdomains with shared sequence similarity, a feature not obvious from the initial sequence examination. Our analysis places this domain into a protein family, named here β-tripod. Using mutagenesis, we identified functionally important regions in the β-tripod domain, which may be involved in target recognition.
Organizational Affiliation:
Agricultural Solutions, BASF, Morrisville, NC 27560; Jelena.zaitseva@agro.basf-se.com.