Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Gray, J.L., von Delft, F., Brennan, P.(2019) Angew Chem Int Ed Engl
- PubMed: 30869179
- DOI: https://doi.org/10.1002/anie.201900585
- Primary Citation of Related Structures:
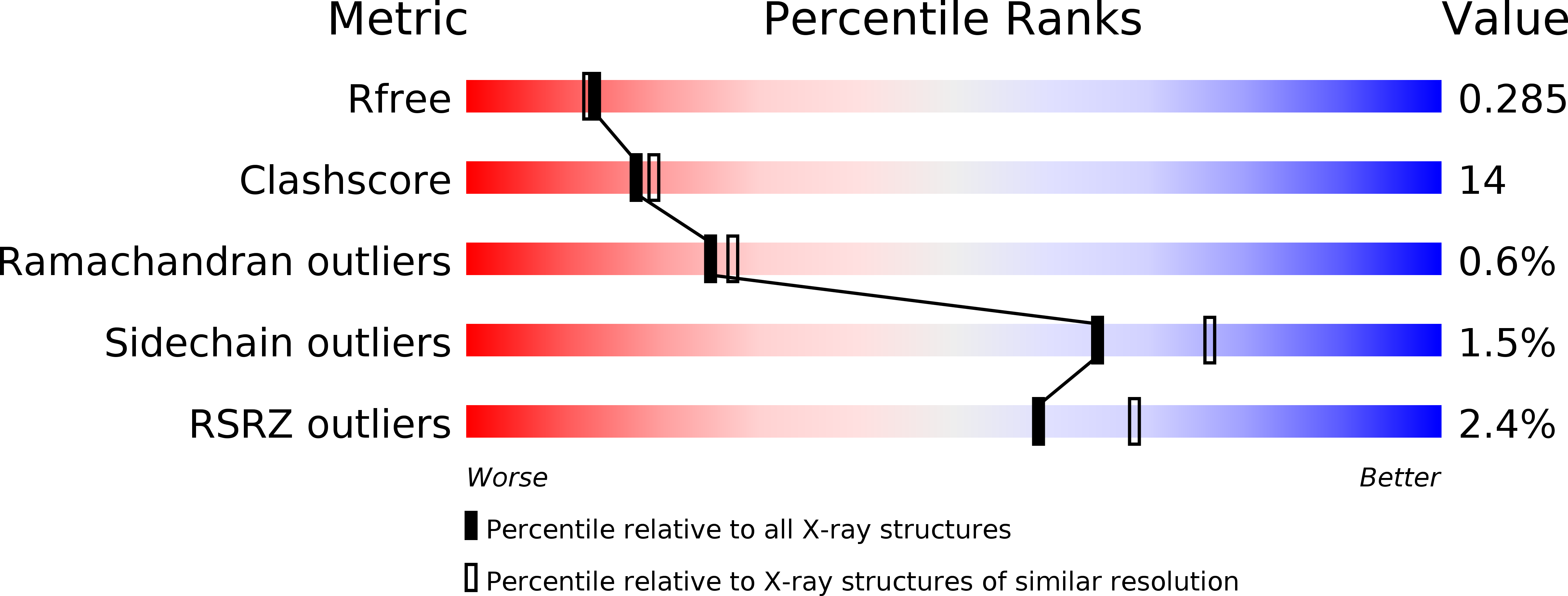
6FAE - PubMed Abstract:
The Ras superfamily of small GTPases are guanine-nucleotide-dependent switches essential for numerous cellular processes. Mutations or dysregulation of these proteins are associated with many diseases, but unsuccessful attempts to target the small GTPases directly have resulted in them being classed as "undruggable". The GTP-dependent signaling of these proteins is controlled by their regulators; guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), and in the Rho and Rab subfamilies, guanine nucleotide dissociation inhibitors (GDIs). This review covers the recent small molecule and biologics strategies to target the small GTPases through their regulators. It seeks to critically re-evaluate recent chemical biology practice, such as the presence of PAINs motifs and the cell-based readout using compounds that are weakly potent or of unknown specificity. It highlights the vast scope of potential approaches for targeting the small GTPases in the future through their regulatory proteins.
Organizational Affiliation:
Structural Genomics Consortium, University of Oxford, NDMRB, Old Road Campus, Oxford, OX3 7DQ, UK.