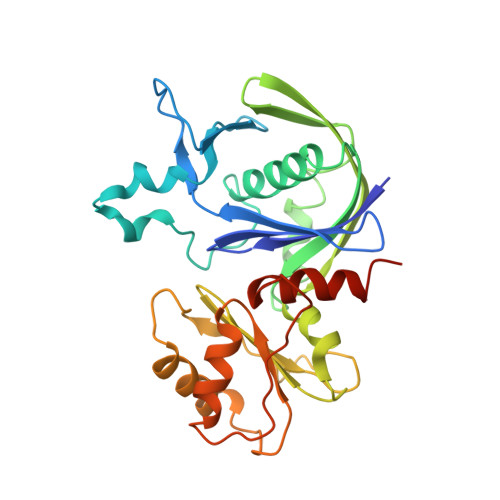
Cryo-EM reconstruction of AlfA fromBacillus subtilisreveals the structure of a simplified actin-like filament at 3.4- angstrom resolution.
Szewczak-Harris, A., Lowe, J.(2018) Proc Natl Acad Sci U S A 115: 3458-3463
- PubMed: 29440489
- DOI: https://doi.org/10.1073/pnas.1716424115
- Primary Citation of Related Structures:
6F95 - PubMed Abstract:
Low copy-number plasmid pLS32 of Bacillus subtilis subsp. natto contains a partitioning system that ensures segregation of plasmid copies during cell division. The partitioning locus comprises actin-like protein AlfA, adaptor protein AlfB, and the centromeric sequence parN Similar to the ParMRC partitioning system from Escherichia coli plasmid R1, AlfA filaments form actin-like double helical filaments that arrange into an antiparallel bipolar spindle, which attaches its growing ends to sister plasmids through interactions with AlfB and parN Because, compared with ParM and other actin-like proteins, AlfA is highly diverged in sequence, we determined the atomic structure of nonbundling AlfA filaments to 3.4-Å resolution by cryo-EM. The structure reveals how the deletion of subdomain IIB of the canonical actin fold has been accommodated by unique longitudinal and lateral contacts, while still enabling formation of left-handed, double helical, polar and staggered filaments that are architecturally similar to ParM. Through cryo-EM reconstruction of bundling AlfA filaments, we obtained a pseudoatomic model of AlfA doublets: the assembly of two filaments. The filaments are antiparallel, as required by the segregation mechanism, and exactly antiphasic with near eightfold helical symmetry, to enable efficient doublet formation. The structure of AlfA filaments and doublets shows, in atomic detail, how deletion of an entire domain of the actin fold is compensated by changes to all interfaces so that the required properties of polymerization, nucleotide hydrolysis, and antiparallel doublet formation are retained to fulfill the system's biological raison d'être.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom.