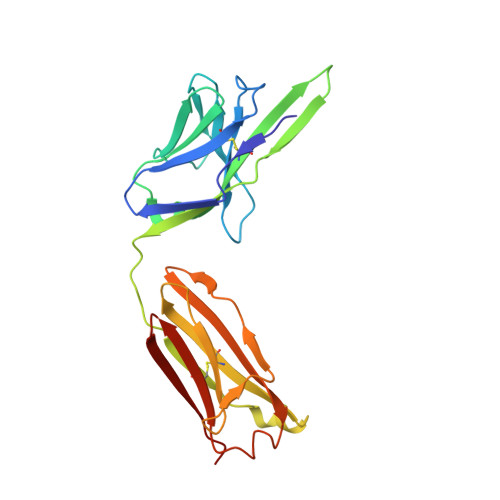
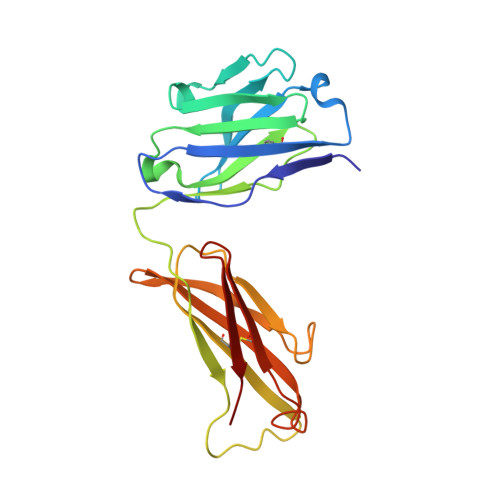
Dissociation of the Dimer of the Intrinsically Disordered Domain of RNase Y upon Antibody Binding.
Hardouin, P., Velours, C., Bou-Nader, C., Assrir, N., Laalami, S., Putzer, H., Durand, D., Golinelli-Pimpaneau, B.(2018) Biophys J 115: 2102-2113
- PubMed: 30447990
- DOI: https://doi.org/10.1016/j.bpj.2018.10.016
- Primary Citation of Related Structures:
6F7T - PubMed Abstract:
Although RNase Y acts as the key enzyme initiating messenger RNA decay in Bacillus subtilis and likely in many other Gram-positive bacteria, its three-dimensional structure remains unknown. An antibody belonging to the rare immunoglobulin G (IgG) 2b λx isotype was raised against a 12-residue conserved peptide from the N-terminal noncatalytic domain of B. subtilis RNase Y (BsRNaseY) that is predicted to be intrinsically disordered. Here, we show that this domain can be produced as a stand-alone protein called Nter-BsRNaseY that undergoes conformational changes between monomeric and dimeric forms. Circular dichroism and size exclusion chromatography coupled with multiangle light scattering or with small angle x-ray scattering indicate that the Nter-BsRNaseY dimer displays an elongated form and a high content of α-helices, in agreement with the existence of a central coiled-coil structure appended with flexible ends, and that the monomeric state of Nter-BsRNaseY is favored upon binding the fragment antigen binding (Fab) of the antibody. The dissociation constants of the IgG/BsRNaseY, IgG/Nter-BsRNaseY, and IgG/peptide complexes indicate that the affinity of the IgG for Nter-BsRNaseY is in the nM range and suggest that the peptide is less accessible in BsRNaseY than in Nter-BsRNaseY. The crystal structure of the Fab in complex with the peptide antigen shows that the peptide adopts an elongated U-shaped conformation in which the unique hydrophobic residue of the peptide, Leu6, is completely buried. The peptide/Fab complex may mimic the interaction of a microdomain of the N-terminal domain of BsRNaseY with one of its cellular partners within the degradosome complex. Altogether, our results suggest that BsRNaseY may become accessible for protein interaction upon dissociation of its N-terminal domain into the monomeric form.
Organizational Affiliation:
Laboratoire de Chimie des Processus Biologiques, UMR 8229 CNRS, Collège de France, Université Pierre et Marie Curie, Paris CEDEX 05, France.