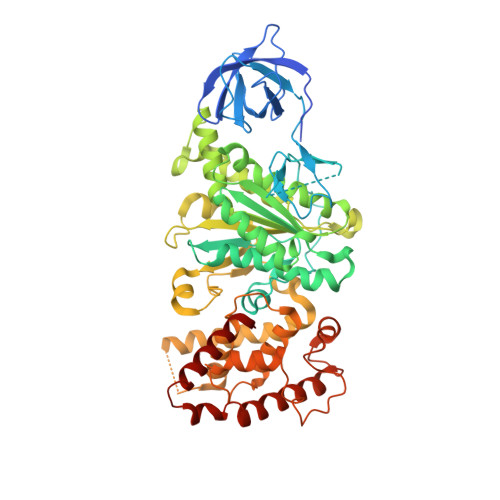
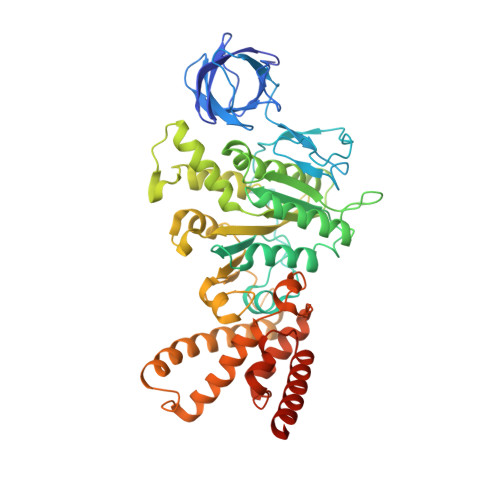
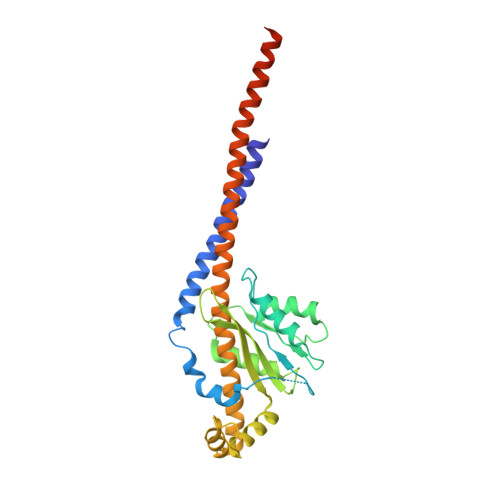
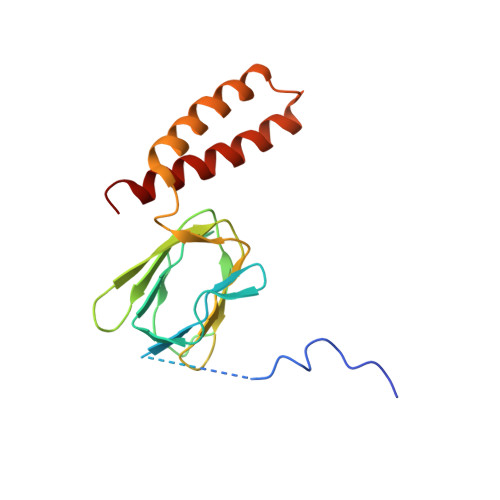
ATP synthase fromTrypanosoma bruceihas an elaborated canonical F1-domain and conventional catalytic sites.
Montgomery, M.G., Gahura, O., Leslie, A.G.W., Zikova, A., Walker, J.E.(2018) Proc Natl Acad Sci U S A 115: 2102-2107
- PubMed: 29440423
- DOI: https://doi.org/10.1073/pnas.1720940115
- Primary Citation of Related Structures:
6F5D - PubMed Abstract:
The structures and functions of the components of ATP synthases, especially those subunits involved directly in the catalytic formation of ATP, are widely conserved in metazoans, fungi, eubacteria, and plant chloroplasts. On the basis of a map at 32.5-Å resolution determined in situ in the mitochondria of Trypanosoma brucei by electron cryotomography, it has been proposed that the ATP synthase in this species has a noncanonical structure and different catalytic sites in which the catalytically essential arginine finger is provided not by the α-subunit adjacent to the catalytic nucleotide-binding site as in all species investigated to date, but rather by a protein, p18, found only in the euglenozoa. A crystal structure at 3.2-Å resolution of the catalytic domain of the same enzyme demonstrates that this proposal is incorrect. In many respects, the structure is similar to the structures of F 1 -ATPases determined previously. The α 3 β 3 -spherical portion of the catalytic domain in which the three catalytic sites are found, plus the central stalk, are highly conserved, and the arginine finger is provided conventionally by the α-subunits adjacent to each of the three catalytic sites found in the β-subunits. Thus, the enzyme has a conventional catalytic mechanism. The structure differs from previous described structures by the presence of a p18 subunit, identified only in the euglenozoa, associated with the external surface of each of the three α-subunits, thereby elaborating the F 1 -domain. Subunit p18 is a pentatricopeptide repeat (PPR) protein with three PPRs and appears to have no function in the catalytic mechanism of the enzyme.
Organizational Affiliation:
The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Cambridge CB2 0XY, United Kingdom.