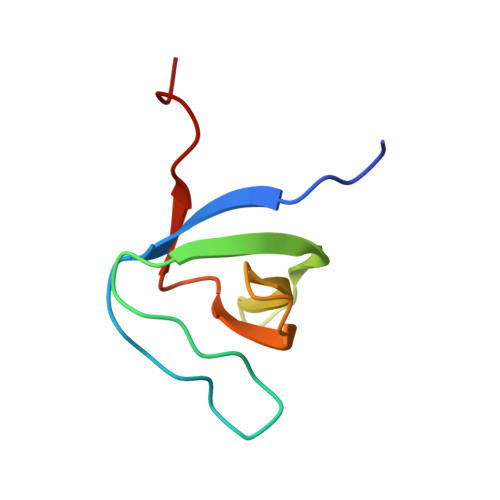
Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Goretzki, B., Glogowski, N.A., Diehl, E., Duchardt-Ferner, E., Hacker, C., Gaudet, R., Hellmich, U.A.(2018) Structure 26: 1583-1593.e5
- PubMed: 30244966
- DOI: https://doi.org/10.1016/j.str.2018.08.002
- Primary Citation of Related Structures:
6F55 - PubMed Abstract:
Transient receptor potential (TRP) channels are polymodally regulated ion channels. TRPV4 (vanilloid 4) is sensitized by PIP 2 and desensitized by Syndapin3/PACSIN3, which bind to the structurally uncharacterized TRPV4 N terminus. We determined the nuclear magnetic resonance structure of the Syndapin3/PACSIN3 SH3 domain in complex with the TRPV4 N-terminal proline-rich region (PRR), which binds as a class I polyproline II (PPII) helix. This PPII conformation is broken by a conserved proline in a cis conformation. Beyond the PPII, we find that the proximal TRPV4 N terminus is unstructured, a feature conserved across species thus explaining the difficulties in resolving it in previous structural studies. Syndapin/PACSIN SH3 domain binding leads to rigidification of both the PRR and the adjacent PIP 2 binding site. We determined the affinities of the TRPV4 N terminus for PACSIN1, 2, and 3 SH3 domains and PIP 2 and deduce a hierarchical interaction network where Syndapin/PACSIN binding influences the PIP 2 binding site but not vice versa.
Organizational Affiliation:
Institute for Pharmacy and Biochemistry, Johannes Gutenberg-Universität Mainz, 55128 Mainz, Germany; Center for Biomolecular Magnetic Resonance (BMRZ), Goethe-Universität, 60438 Frankfurt am Main, Germany.