Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Klusch, N., Murphy, B.J., Mills, D.J., Yildiz, O., Kuhlbrandt, W.(2017) Elife 6
- PubMed: 29210357
- DOI: https://doi.org/10.7554/eLife.33274
- Primary Citation of Related Structures:
6F36 - PubMed Abstract:
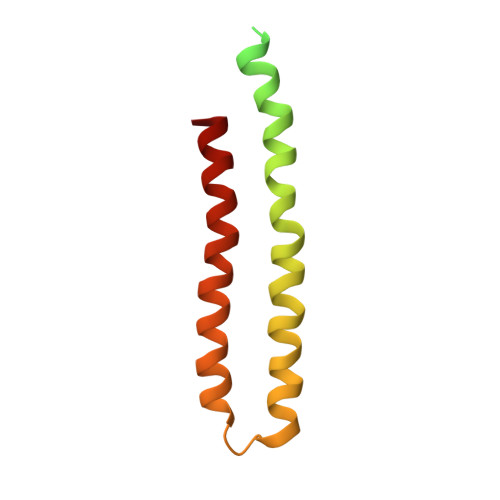
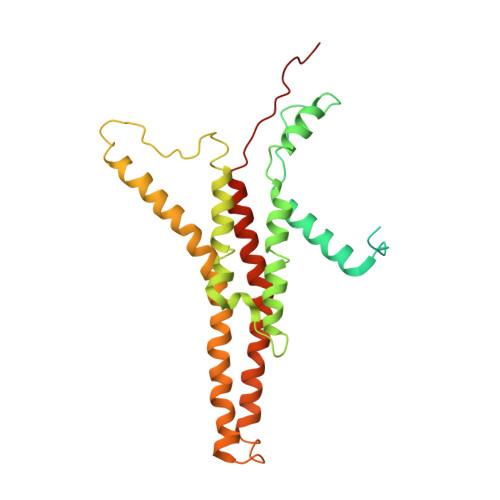
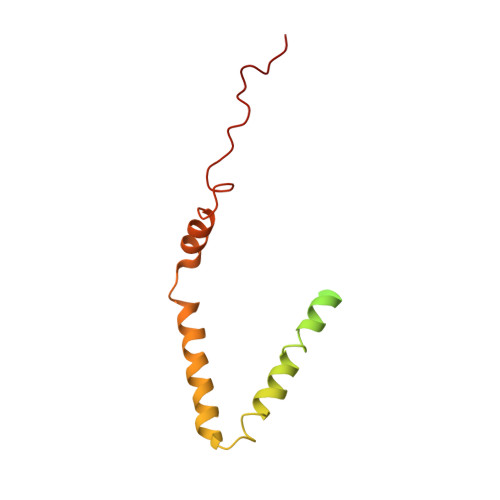
ATP synthases produce ATP by rotary catalysis, powered by the electrochemical proton gradient across the membrane. Understanding this fundamental process requires an atomic model of the proton pathway. We determined the structure of an intact mitochondrial ATP synthase dimer by electron cryo-microscopy at near-atomic resolution. Charged and polar residues of the a -subunit stator define two aqueous channels, each spanning one half of the membrane. Passing through a conserved membrane-intrinsic helix hairpin, the lumenal channel protonates an acidic glutamate in the c -ring rotor. Upon ring rotation, the protonated glutamate encounters the matrix channel and deprotonates. An arginine between the two channels prevents proton leakage. The steep potential gradient over the sub-nm inter-channel distance exerts a force on the deprotonated glutamate, resulting in net directional rotation.
Organizational Affiliation:
Department of Structural Biology, Max Planck Institute of Biophysics, Frankfurt, Germany.