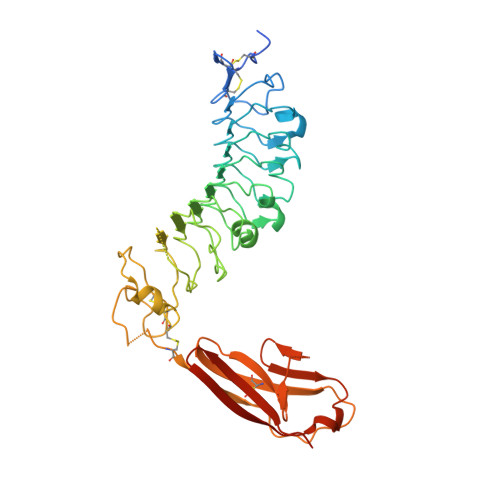
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Karki, S., Paudel, P., Sele, C., Shkumatov, A.V., Kajander, T.(2018) Protein Eng Des Sel 31: 147-157
- PubMed: 29897575
- DOI: https://doi.org/10.1093/protein/gzy012
- Primary Citation of Related Structures:
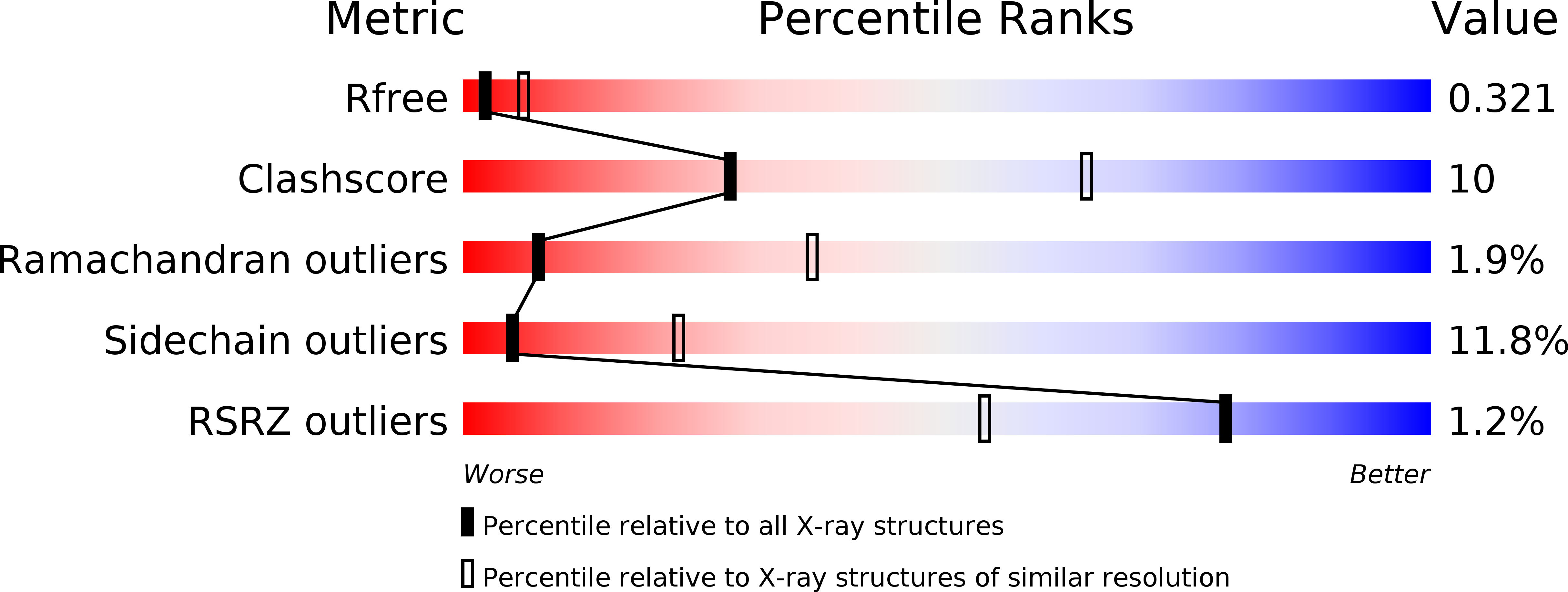
6F2O - PubMed Abstract:
Synaptic adhesion molecules play a crucial role in the regulation of synapse development and maintenance. Recently, several families of leucine-rich repeat (LRR) domain-containing neuronal adhesion molecules have been characterised, including netrin-G ligands, LRRTMs and the synaptic adhesion-like molecule (SALM) family proteins. Most of these are expressed at the excitatory glutamatergic synapses, and dysfunctions of these genes are genetically linked with cognitive disorders, such as autism spectrum disorders and schizophrenia. The SALM family proteins SALM3 and SALM5, similar to SLITRKs, have been shown to bind to the presynaptic receptor protein tyrosine phosphatase (RPTP) family ligands. Here, we present the 3.1 Å crystal structure of the SALM5 LRR-Ig-domain construct and biophysical studies that verify the crystallographic results. We show that SALM1, SALM3 and SALM5 form similar dimeric structures, in which the LRR domains form the dimer interface. Both SALM3 and SALM5 bind to RPTP immunoglobulin domains with micromolar affinity. SALM3 shows a clear preference for the RPTP ligands with the meB splice insert. Our structural studies and sequence conservation analysis suggests a ligand-binding site and mechanism for RPTP binding via the dimeric LRR domain region.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, Helsinki 00014, Finland.