Interferons type II and their receptors R1 and R2 in fish species: Evolution, structure, and function.
Zahradnik, J., Kolarova, L., Parizkova, H., Kolenko, P., Schneider, B.(2018) Fish Shellfish Immunol 79: 140-152
- PubMed: 29742458
- DOI: https://doi.org/10.1016/j.fsi.2018.05.008
- Primary Citation of Related Structures:
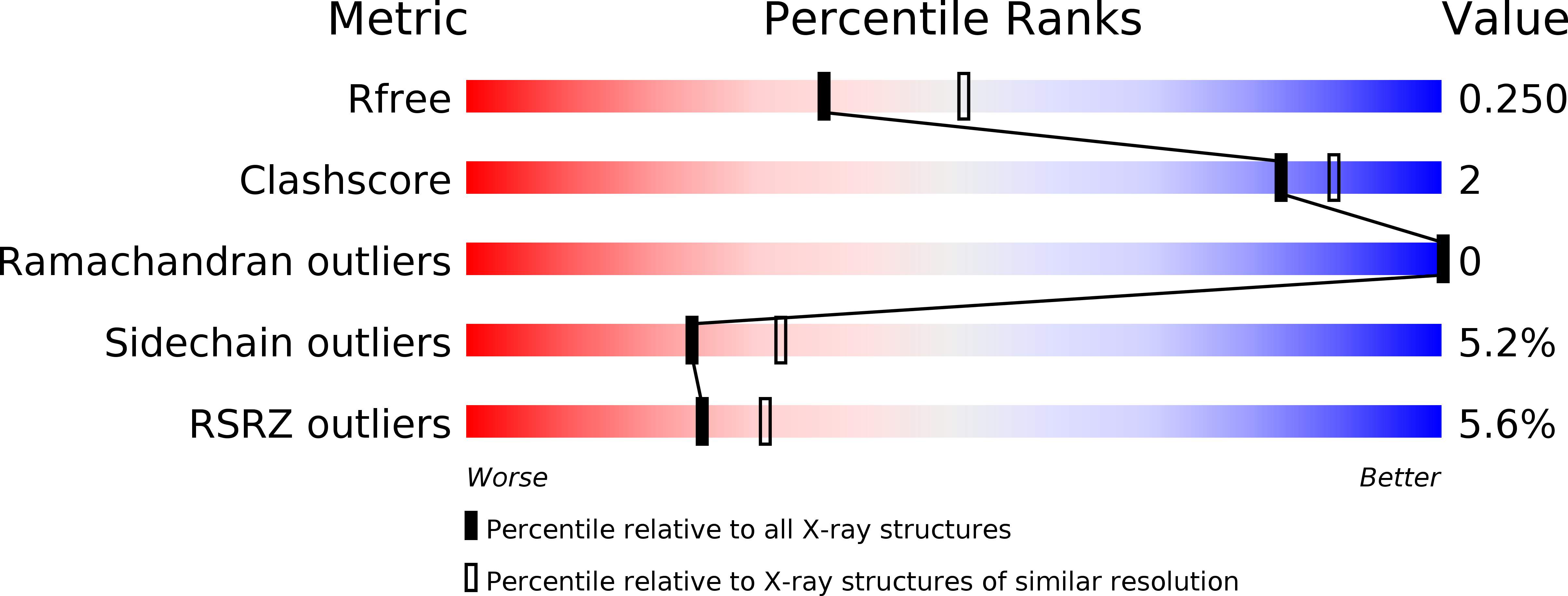
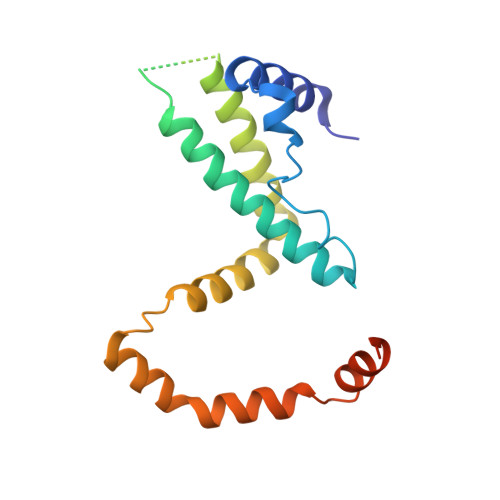
6F1E - PubMed Abstract:
Interferon gamma (IFN-γ) is one of the key players in the immune system of vertebrates. The evolution and properties of IFN-γ and its receptors in fish species are of special interest as they point to the origin of innate immunity in vertebrates. We studied the phylogeny, biophysical and structural properties of IFN-γ and its receptors. Our phylogeny analysis suggests the existence of two groups of IFN-γ related proteins, one specific for Acanthomorpha, the other for Cypriniformes, Characiformes and Siluriformes. The analysis further shows an ancient duplication of the gene for IFN-γ receptor 1 (IFN- γR1) and the parallel existence of the duplicated genes in all current teleost fish species. In contrast, only one gene can be found for receptor 2, IFN- γR2. The specificity of the interaction between IFN- γ and both types of IFN- γR1 was determined by microscale thermophoresis measurements of the equilibrium dissociation constants for the proteins from three fish species. The measured preference of IFN- γ for one of the two forms of receptor 1agrees with the bioinformatic analysis of the coevolution between IFN- γ and receptor 1. To elucidate structural relationships between IFN-γ of fish and other vertebrate species, we determined the crystal structure of IFN-γ from olive flounder (Paralichthys olivaceus, PoliIFN-γ) at crystallographic resolution of 2.3 Å and the low-resolution structures of Takifugu rubripes, Oreochromis niloticus, and Larimichthys crocea IFN-γ by small angle X-ray diffraction. The overall PoliIFN-γ fold is the same as the fold of the other known IFN- γ structures but there are some significant structural differences, namely the additional C-terminal helix G and a different angle between helices C and D in PoliIFN-γ.
Organizational Affiliation:
Laboratory of Biomolecular Recognition, Institute of Biotechnology of the Czech Academy of Sciences, v. v. i., BIOCEV, Průmyslová 595, CZ-252 42 Vestec, Czech Republic. Electronic address: jiri.zahradnik@ibt.cas.cz.