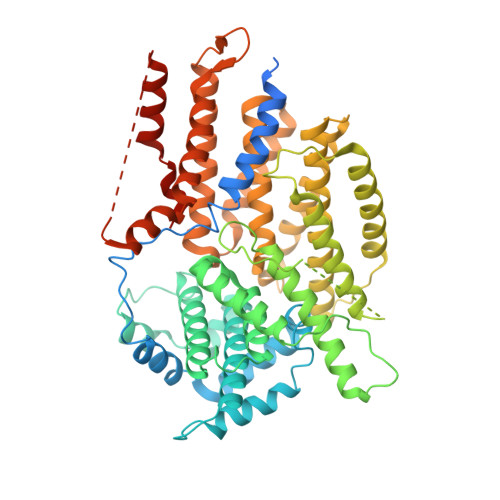
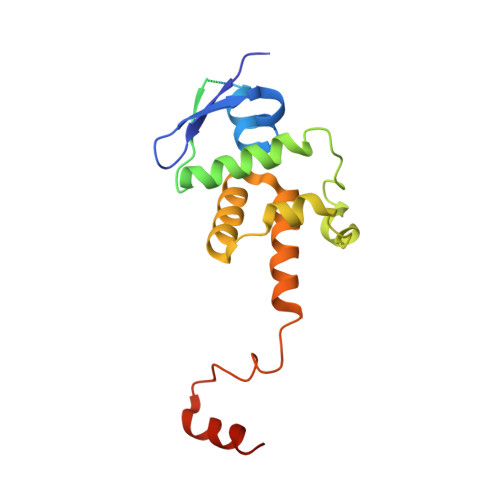
Structural basis for assembly of the CBF3 kinetochore complex.
Leber, V., Nans, A., Singleton, M.R.(2018) EMBO J 37: 269-281
- PubMed: 29212814
- DOI: https://doi.org/10.15252/embj.201798134
- Primary Citation of Related Structures:
6F07 - PubMed Abstract:
Eukaryotic chromosomes contain a specialised region known as the centromere, which forms the platform for kinetochore assembly and microtubule attachment. The centromere is distinguished by the presence of nucleosomes containing the histone H3 variant, CENP-A. In budding yeast, centromere establishment begins with the recognition of a specific DNA sequence by the CBF3 complex. This in turn facilitates CENP-A Cse4 nucleosome deposition and kinetochore assembly. Here, we describe a 3.6 Å single-particle cryo-EM reconstruction of the core CBF3 complex, incorporating the sequence-specific DNA-binding protein Cep3 together with regulatory subunits Ctf13 and Skp1. This provides the first structural data on Ctf13, defining it as an F-box protein of the leucine-rich-repeat family, and demonstrates how a novel F-box-mediated interaction between Ctf13 and Skp1 is responsible for initial assembly of the CBF3 complex.
Organizational Affiliation:
Structural Biology of Chromosome Segregation Laboratory, The Francis Crick Institute, London, UK.