Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity.
Celma, L., Corbinais, C., Vercruyssen, J., Veaute, X., de la Sierra-Gallay, I.L., Guerois, R., Busso, D., Mathieu, A., Marsin, S., Quevillon-Cheruel, S., Radicella, J.P.(2017) PLoS One 12: e0189049
- PubMed: 29206236
- DOI: https://doi.org/10.1371/journal.pone.0189049
- Primary Citation of Related Structures:
6EHI - PubMed Abstract:
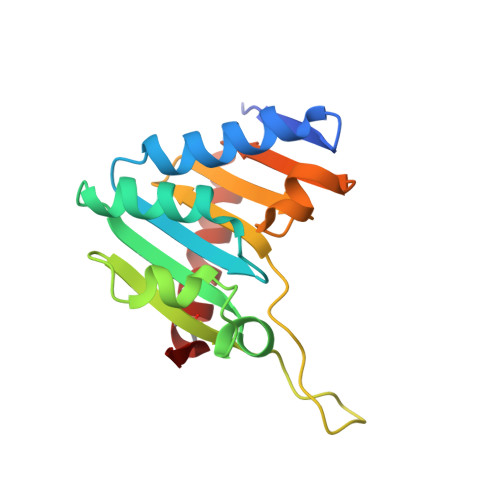
The Phospholipase D (PLD) superfamily of proteins includes a group of enzymes with nuclease activity on various nucleic acid substrates. Here, with the aim of better understanding the substrate specificity determinants in this subfamily, we have characterised the enzymatic activity and the crystal structure of NucT, a nuclease implicated in Helicobacter pylori purine salvage and natural transformation and compared them to those of its bacterial and mammalian homologues. NucT exhibits an endonuclease activity with a strong preference for single stranded nucleic acids substrates. We identified histidine124 as essential for the catalytic activity of the protein. Comparison of the NucT crystal structure at 1.58 Å resolution reported here with those of other members of the sub-family suggests that the specificity of NucT for single-stranded nucleic acids is provided by the width of a positively charged groove giving access to the catalytic site.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Université Paris-Sud, Gif-sur-Yvette cedex, France.