Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Wang, F., Gu, Y., O'Brien, J.P., Yi, S.M., Yalcin, S.E., Srikanth, V., Shen, C., Vu, D., Ing, N.L., Hochbaum, A.I., Egelman, E.H., Malvankar, N.S.(2019) Cell 177: 361-369.e10
- PubMed: 30951668
- DOI: https://doi.org/10.1016/j.cell.2019.03.029
- Primary Citation of Related Structures:
6EF8 - PubMed Abstract:
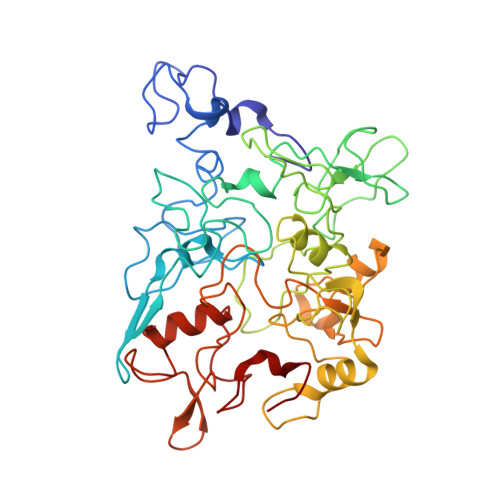
Long-range (>10 μm) transport of electrons along networks of Geobacter sulfurreducens protein filaments, known as microbial nanowires, has been invoked to explain a wide range of globally important redox phenomena. These nanowires were previously thought to be type IV pili composed of PilA protein. Here, we report a 3.7 Å resolution cryoelectron microscopy structure, which surprisingly reveals that, rather than PilA, G. sulfurreducens nanowires are assembled by micrometer-long polymerization of the hexaheme cytochrome OmcS, with hemes packed within ∼3.5-6 Å of each other. The inter-subunit interfaces show unique structural elements such as inter-subunit parallel-stacked hemes and axial coordination of heme by histidines from neighboring subunits. Wild-type OmcS filaments show 100-fold greater conductivity than other filaments from a ΔomcS strain, highlighting the importance of OmcS to conductivity in these nanowires. This structure explains the remarkable capacity of soil bacteria to transport electrons to remote electron acceptors for respiration and energy sharing.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA 22908, USA.