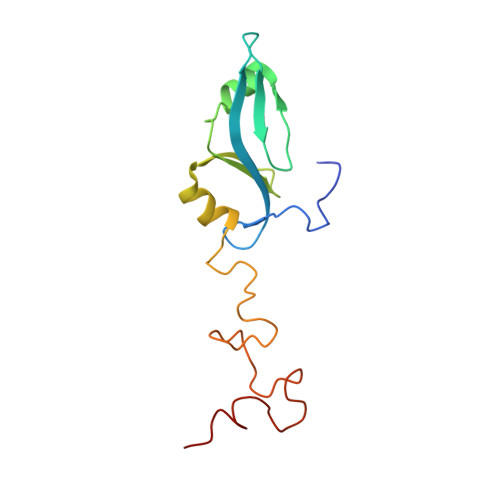
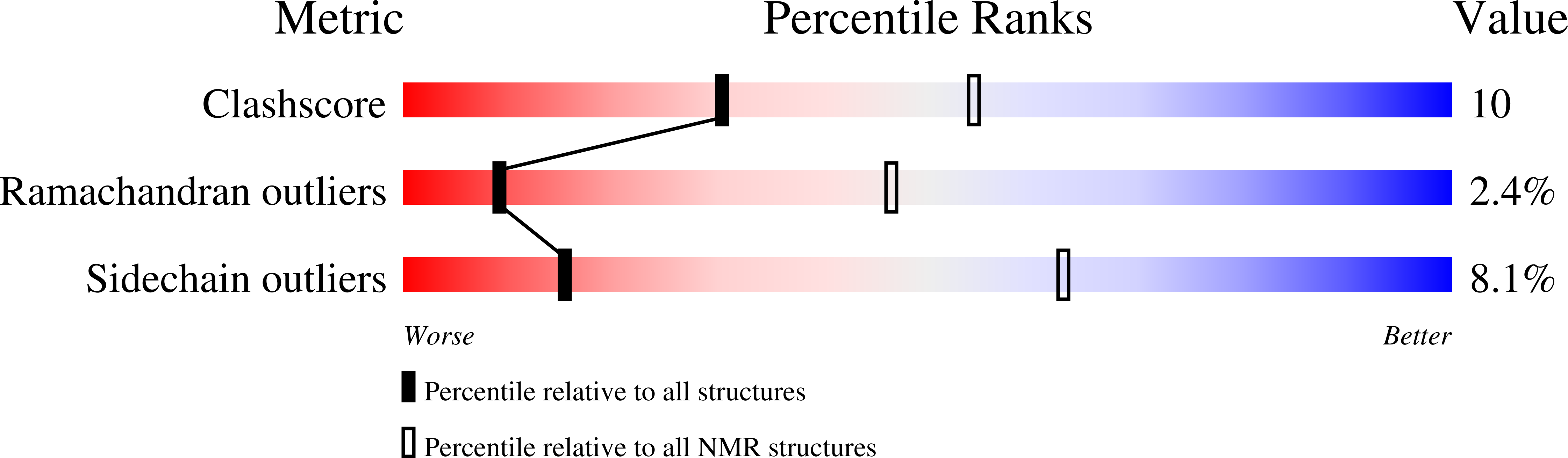The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy.
Newcomer, R.L., Schrad, J.R., Gilcrease, E.B., Casjens, S.R., Feig, M., Teschke, C.M., Alexandrescu, A.T., Parent, K.N.(2019) Elife 8
- PubMed: 30945633
- DOI: https://doi.org/10.7554/eLife.45345
- Primary Citation of Related Structures:
6E3C - PubMed Abstract:
The major coat proteins of dsDNA tailed phages (order Caudovirales ) and herpesviruses form capsids by a mechanism that includes active packaging of the dsDNA genome into a precursor procapsid, followed by expansion and stabilization of the capsid. These viruses have evolved diverse strategies to fortify their capsids, such as non-covalent binding of auxiliary 'decoration' (Dec) proteins. The Dec protein from the P22-like phage L has a highly unusual binding strategy that distinguishes between nearly identical three-fold and quasi-three-fold sites of the icosahedral capsid. Cryo-electron microscopy and three-dimensional image reconstruction were employed to determine the structure of native phage L particles. NMR was used to determine the structure/dynamics of Dec in solution. The NMR structure and the cryo-EM density envelope were combined to build a model of the capsid-bound Dec trimer. Key regions that modulate the binding interface were verified by site-directed mutagenesis.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of Connecticut, Storrs, United States.