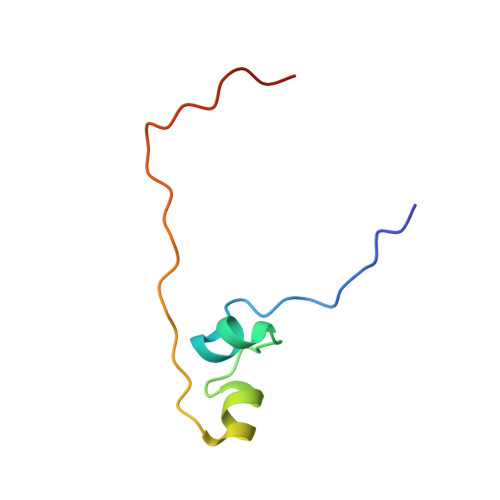
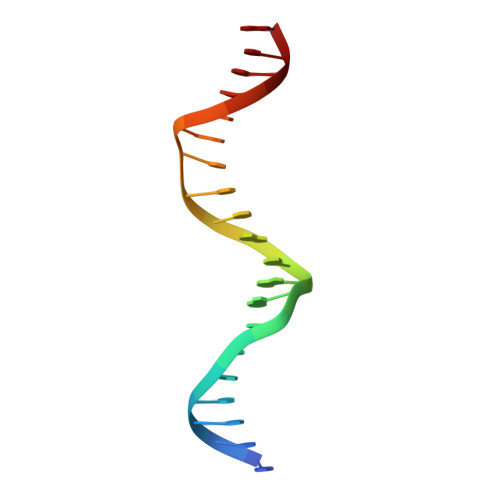
Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Garg, A., Goldgur, Y., Schwer, B., Shuman, S.(2018) Nucleic Acids Res 46: 11262-11273
- PubMed: 30212894
- DOI: https://doi.org/10.1093/nar/gky827
- Primary Citation of Related Structures:
6E33 - PubMed Abstract:
Pho7, a member of the Zn2Cys6 family of fungal transcription factors, is the key transcriptional activator underlying fission yeast phosphate homeostasis, a physiological response to phosphate starvation in which the pho1, pho84 and tgp1 genes are upregulated. Here, we delineated a minimized 61-amino-acid Pho7 DNA-binding domain (DBD) and determined the 1.7 Å crystal structure of the DBD at its target site in the tgp1 promoter. Two distinctive features of the Pho7 DBD are: it binds DNA as a monomer, unlike most other fungal zinc-cluster factors that bind as homodimers; and it makes extensive interactions with its asymmetric target sequence over a 14-bp footprint that entails hydrogen bonding to 13 individual bases within, and remote from, the CGG triplet typically recognized by other Zn2Cys6 DBDs. Base pair substitutions at Pho7 sites in the tgp1 and pho1 promoters highlight the importance of the 5'-CGG triplet for Pho7 binding in vitro and Pho7-dependent gene expression in vivo. We identify several DBD amino acids at which alanine substitution effaced or attenuated the pho1 phosphate starvation response and concordantly reduced Pho7 binding to a pho1 promoter site.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.