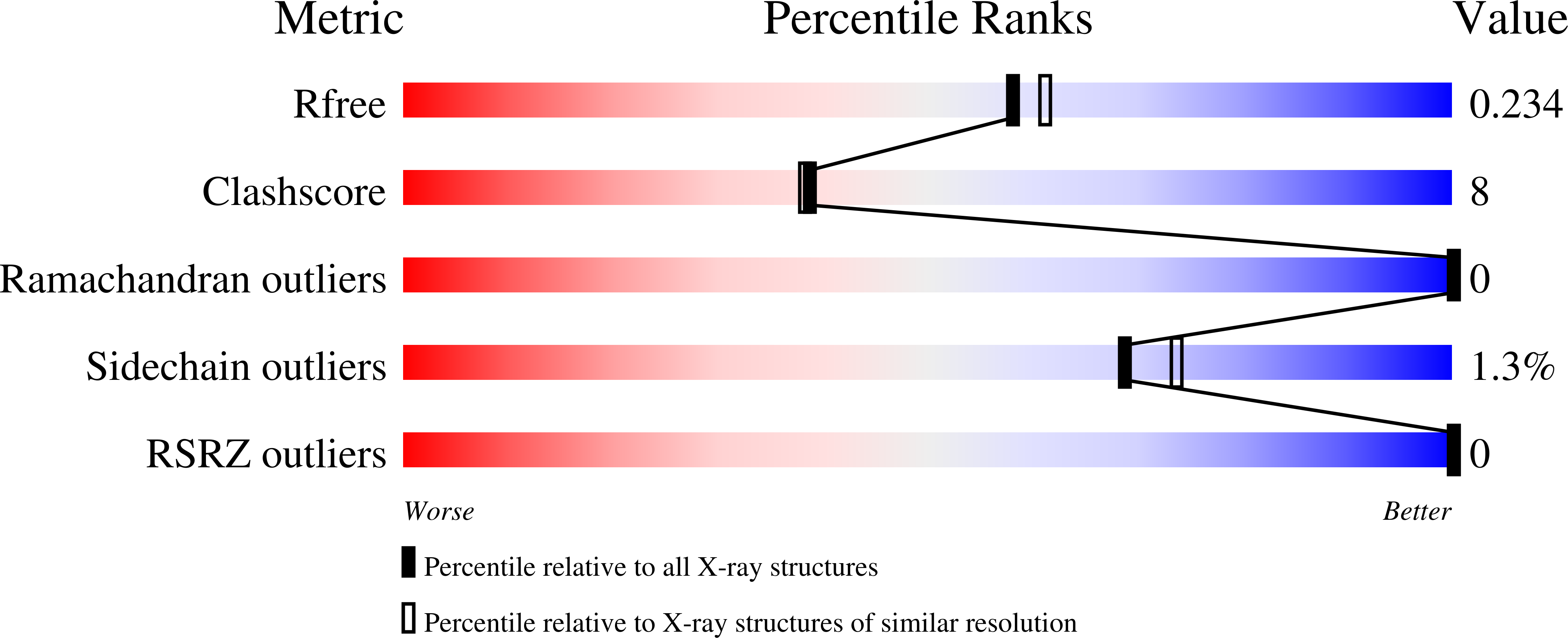
Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Ghosh, A., Banerjee, A., Amzel, L.M., Vasta, G.R., Bianchet, M.A.(2019) Glycobiology 29: 419-430
- PubMed: 30834446
- DOI: https://doi.org/10.1093/glycob/cwz015
- Primary Citation of Related Structures:
6E20 - PubMed Abstract:
Galectins, highly conserved β-galactoside-binding lectins, have diverse regulatory roles in development and immune homeostasis and can mediate protective functions during microbial infection. In recent years, the role of galectins in viral infection has generated considerable interest. Studies on highly pathogenic viruses have provided invaluable insight into the participation of galectins in various stages of viral infection, including attachment and entry. Detailed mechanistic and structural aspects of these processes remain undetermined. To address some of these gaps in knowledge, we used Zebrafish as a model system to examine the role of galectins in infection by infectious hematopoietic necrosis virus (IHNV), a rhabdovirus that is responsible for significant losses in both farmed and wild salmonid fish. Like other rhabdoviruses, IHNV is characterized by an envelope consisting of trimers of a glycoprotein that display multiple N-linked oligosaccharides and play an integral role in viral infection by mediating the virus attachment and fusion. Zebrafish's proto-typical galectin Drgal1-L2 and the chimeric-type galectin Drgal3-L1 interact directly with the glycosylated envelope of IHNV, and significantly reduce viral attachment. In this study, we report the structure of the complex of Drgal1-L2 with N-acetyl-d-lactosamine at 2.0 Å resolution. To gain structural insight into the inhibitory effect of these galectins on IHNV attachment to the zebrafish epithelial cells, we modeled Drgal3-L1 based on human galectin-3, as well as, the ectodomain of the IHNV glycoprotein. These models suggest mechanisms for which the binding of these galectins to the IHNV glycoprotein hinders with different potencies the viral attachment required for infection.
Organizational Affiliation:
Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA.