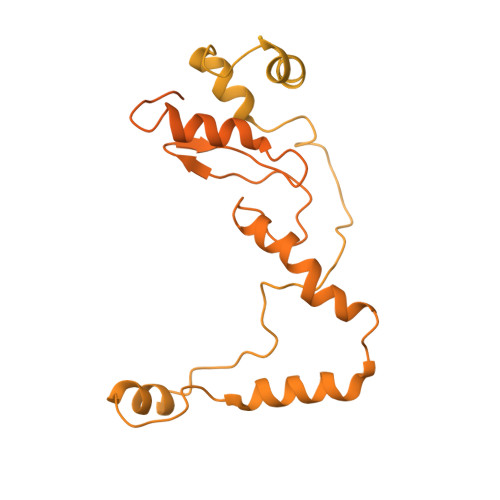
Malaria parasite translocon structure and mechanism of effector export.
Ho, C.M., Beck, J.R., Lai, M., Cui, Y., Goldberg, D.E., Egea, P.F., Zhou, Z.H.(2018) Nature 561: 70-75
- PubMed: 30150771
- DOI: https://doi.org/10.1038/s41586-018-0469-4
- Primary Citation of Related Structures:
6E10, 6E11 - PubMed Abstract:
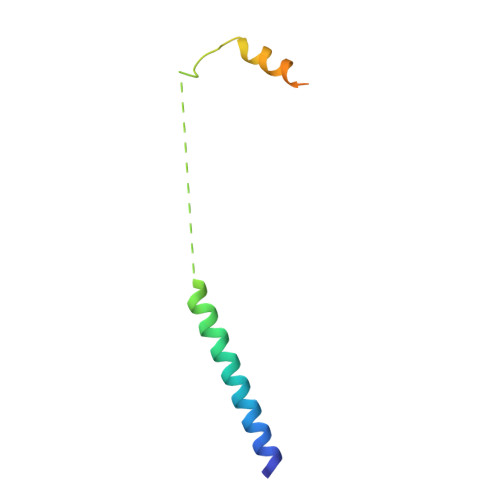
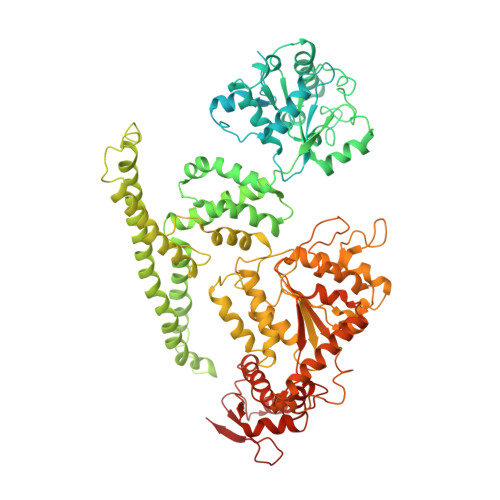
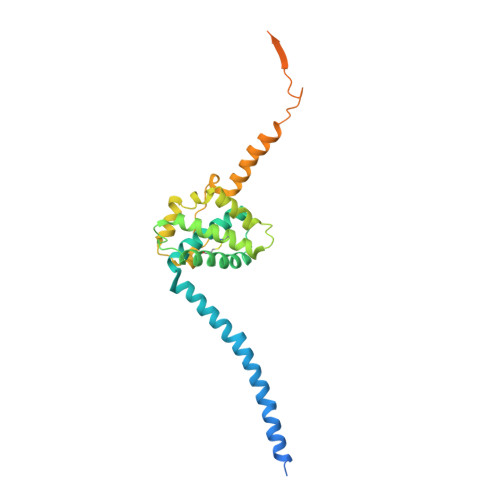
The putative Plasmodium translocon of exported proteins (PTEX) is essential for transport of malarial effector proteins across a parasite-encasing vacuolar membrane into host erythrocytes, but the mechanism of this process remains unknown. Here we show that PTEX is a bona fide translocon by determining structures of the PTEX core complex at near-atomic resolution using cryo-electron microscopy. We isolated the endogenous PTEX core complex containing EXP2, PTEX150 and HSP101 from Plasmodium falciparum in the 'engaged' and 'resetting' states of endogenous cargo translocation using epitope tags inserted using the CRISPR-Cas9 system. In the structures, EXP2 and PTEX150 interdigitate to form a static, funnel-shaped pseudo-seven-fold-symmetric protein-conducting channel spanning the vacuolar membrane. The spiral-shaped AAA+ HSP101 hexamer is tethered above this funnel, and undergoes pronounced compaction that allows three of six tyrosine-bearing pore loops lining the HSP101 channel to dissociate from the cargo, resetting the translocon for the next threading cycle. Our work reveals the mechanism of P. falciparum effector export, and will inform structure-based design of drugs targeting this unique translocon.
Organizational Affiliation:
The Molecular Biology Institute, University of California, Los Angeles, CA, USA.