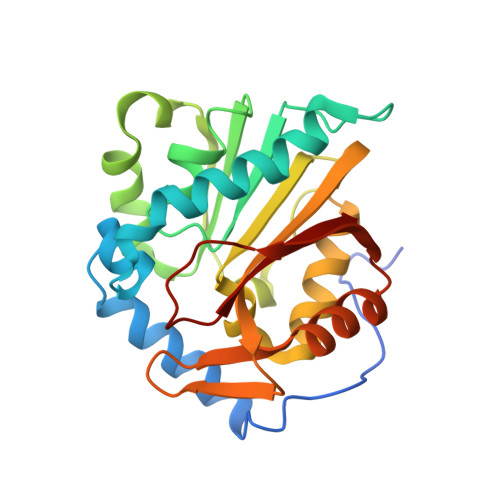
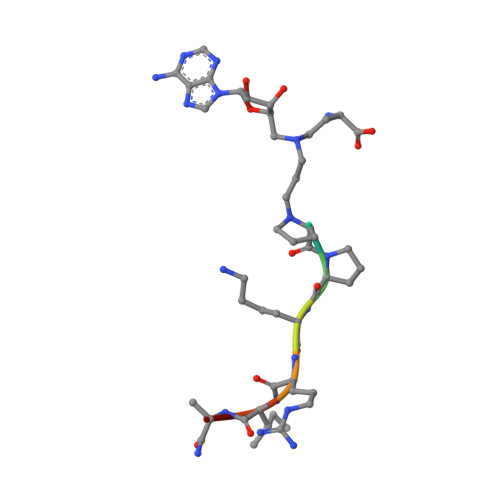
Discovery of Bisubstrate Inhibitors for Protein N-Terminal Methyltransferase 1.
Chen, D., Dong, G., Noinaj, N., Huang, R.(2019) J Med Chem 62: 3773-3779
- PubMed: 30883119
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00206
- Primary Citation of Related Structures:
6DTN - PubMed Abstract:
Protein N-terminal methyltransferase 1 (NTMT1) plays an important role in regulating mitosis and DNA repair. Here, we describe the discovery of a potent NTMT1 bisubstrate inhibitor 4 (IC 50 = 35 ± 2 nM) that exhibits greater than 100-fold selectivity against a panel of methyltransferases. We also report the first crystal structure of NTMT1 in complex with an inhibitor, which revealed that 4 occupies substrate and cofactor binding sites of NTMT1.
Organizational Affiliation:
Department of Medicinal Chemistry and Molecular Pharmacology, Center for Cancer Research, Institute for Drug Discovery , Purdue University , West Lafayette , Indiana 47907 , United States.