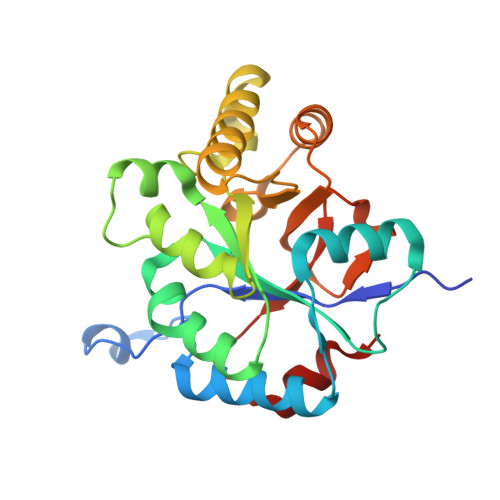
PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Rush, J.S., Parajuli, P., Ruda, A., Li, J., Pohane, A.A., Zamakhaeva, S., Rahman, M.M., Chang, J.C., Gogos, A., Kenner, C.W., Lambeau, G., Federle, M.J., Korotkov, K.V., Widmalm, G., Korotkova, N.(2022) Nat Commun 13: 590