Biophysical insights into a highly selective l-arginine-binding lipoprotein of a pathogenic treponeme.
Deka, R.K., Liu, W.Z., Tso, S.C., Norgard, M.V., Brautigam, C.A.(2018) Protein Sci 27: 2037-2050
- PubMed: 30242931
- DOI: https://doi.org/10.1002/pro.3510
- Primary Citation of Related Structures:
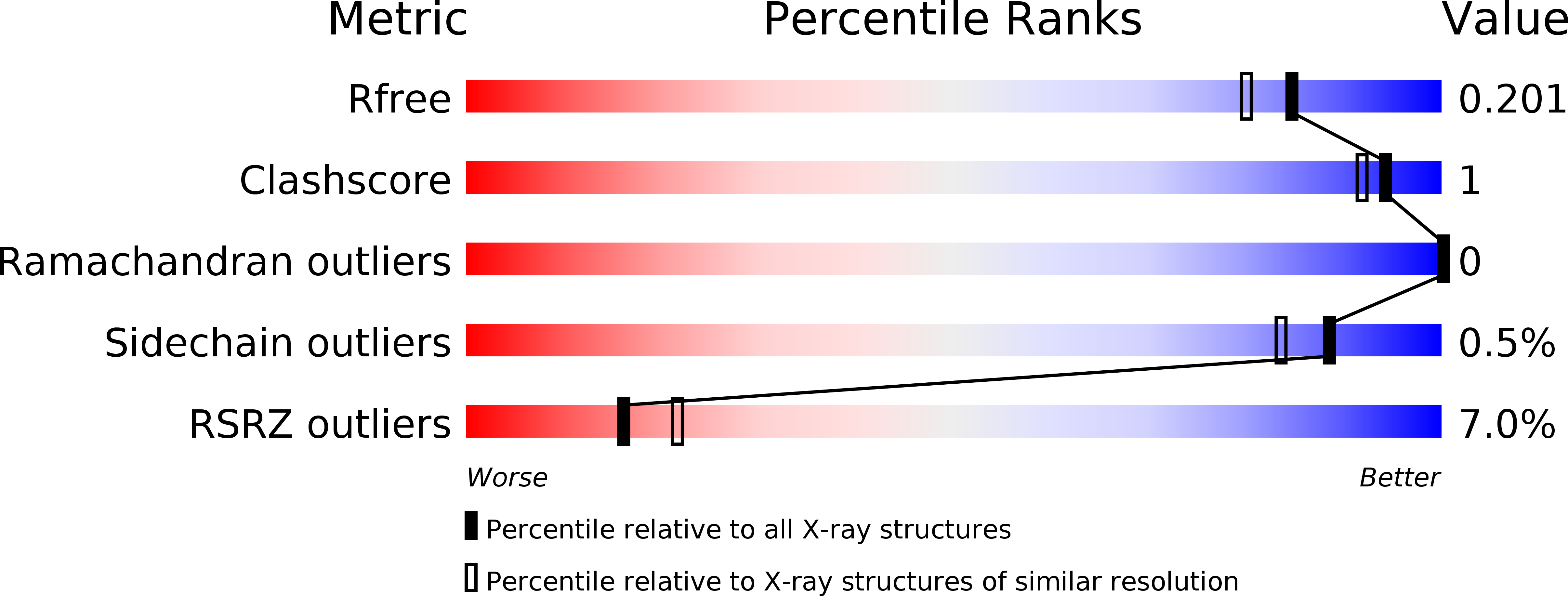
6DET - PubMed Abstract:
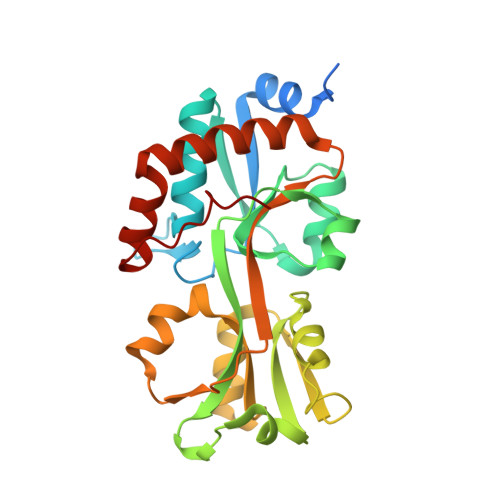
Biophysical and biochemical studies on the lipoproteins and other periplasmic proteins from the spirochetal species Treponema pallidum have yielded numerous insights into the functioning of the organism's peculiar membrane organization, its nutritional requirements, and intermediary metabolism. However, not all T. pallidum proteins have proven to be amenable to biophysical studies. One such recalcitrant protein is Tp0309, a putative polar-amino-acid-binding protein of an ABC transporter system. To gain further information on its possible function, a homolog of the protein from the related species T. vincentii was used as a surrogate. This protein, Tv2483, was crystallized, resulting in the determination of its crystal structure at a resolution of 1.75 Å. The protein has a typical fold for a ligand-binding protein, and a single molecule of l-arginine was bound between its two lobes. Differential scanning fluorimetry and isothermal titration calorimetry experiments confirmed that l-arginine bound to the protein with unusually high selectivity. However, further comparison to Tp0309 showed differences in key amino-acid-binding residues may impart an alternate specificity for the T. pallidum protein.
Organizational Affiliation:
Departments of Microbiology, 5323 Harry Hines Blvd., Dallas, Texas, 75390.