Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Mak, W.S., Wang, X., Arenas, R., Cui, Y., Bertolani, S., Deng, W.Q., Tagkopoulos, I., Wilson, D.K., Siegel, J.B.(2020) Biochemistry 59: 3834-3843
- PubMed: 32935984
- DOI: https://doi.org/10.1021/acs.biochem.0c00665
- Primary Citation of Related Structures:
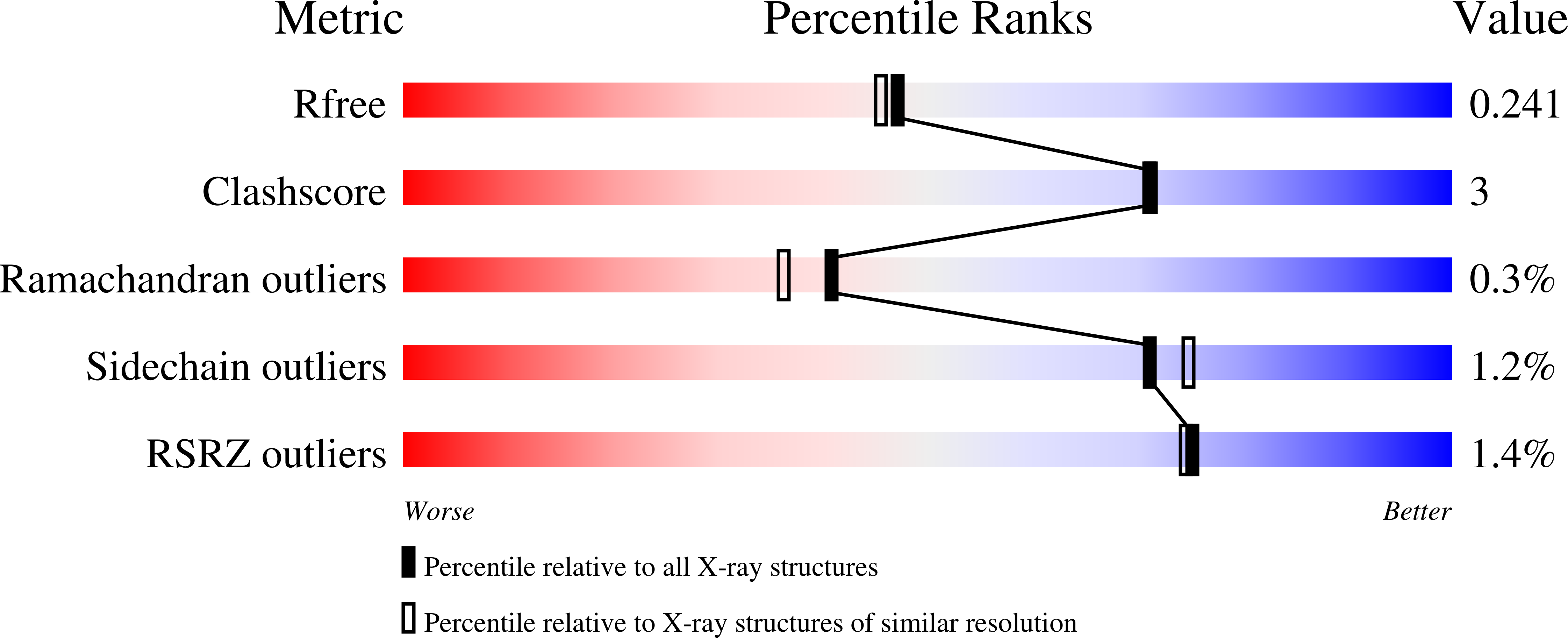
6D9F - PubMed Abstract:
To complement established rational and evolutionary protein design approaches, significant efforts are being made to utilize computational modeling and the diversity of naturally occurring protein sequences. Here, we combine structural biology, genomic mining, and computational modeling to identify structural features critical to aldehyde deformylating oxygenases (ADOs), an enzyme family that has significant implications in synthetic biology and chemoenzymatic synthesis. Through these efforts, we discovered latent ADO-like function across the ferritin-like superfamily in various species of Bacteria and Archaea. We created a machine learning model that uses protein structural features to discriminate ADO-like activity. Computational enzyme design tools were then utilized to introduce ADO-like activity into the small subunit of Escherichia coli class I ribonucleotide reductase. The integrated approach of genomic mining, structural biology, molecular modeling, and machine learning has the potential to be utilized for rapid discovery and modulation of functions across enzyme families.
Organizational Affiliation:
Department of Chemistry, University of California, Davis, One Shields Avenue, Davis, California 95616, United States.