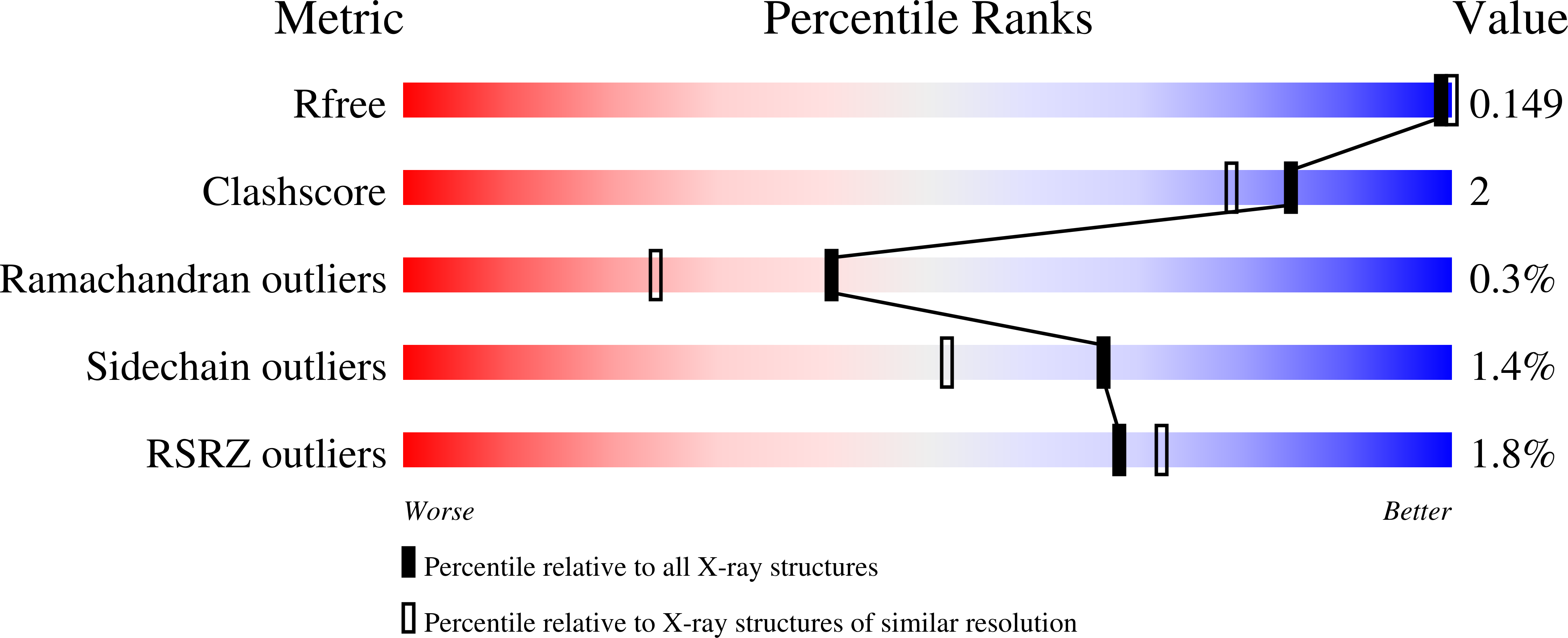
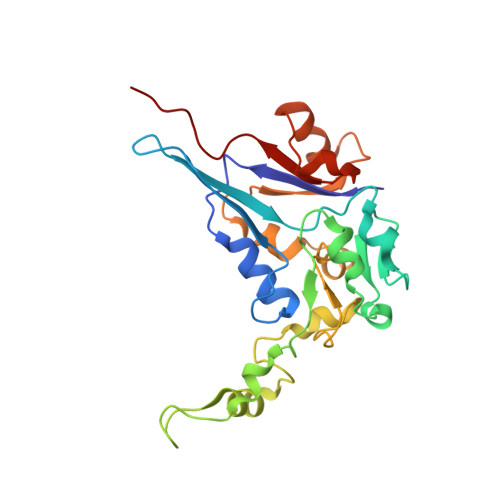
Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Glasser, N.R., Oyala, P.H., Osborne, T.H., Santini, J.M., Newman, D.K.(2018) Proc Natl Acad Sci U S A 115: E8614-E8623
- PubMed: 30104376
- DOI: https://doi.org/10.1073/pnas.1807984115
- Primary Citation of Related Structures:
6CZ7, 6CZ8, 6CZ9, 6CZA - PubMed Abstract:
Arsenate respiration by bacteria was discovered over two decades ago and is catalyzed by diverse organisms using the well-conserved Arr enzyme complex. Until now, the mechanisms underpinning this metabolism have been relatively opaque. Here, we report the structure of an Arr complex (solved by X-ray crystallography to 1.6-Å resolution), which was enabled by an improved Arr expression method in the genetically tractable arsenate respirer Shewanella sp. ANA-3. We also obtained structures bound with the substrate arsenate (1.8 Å), the product arsenite (1.8 Å), and the natural inhibitor phosphate (1.7 Å). The structures reveal a conserved active-site motif that distinguishes Arr [(R/K)GRY] from the closely related arsenite respiratory oxidase (Arx) complex (XGRGWG). Arr activity assays using methyl viologen as the electron donor and arsenate as the electron acceptor display two-site ping-pong kinetics. A Mo(V) species was detected with EPR spectroscopy, which is typical for proteins with a pyranopterin guanine dinucleotide cofactor. Arr is an extraordinarily fast enzyme that approaches the diffusion limit ( K m = 44.6 ± 1.6 μM, k cat = 9,810 ± 220 seconds -1 ), and phosphate is a competitive inhibitor of arsenate reduction ( K i = 325 ± 12 μM). These observations, combined with knowledge of typical sedimentary arsenate and phosphate concentrations and known rates of arsenate desorption from minerals in the presence of phosphate, suggest that ( i ) arsenate desorption limits microbiologically induced arsenate reductive mobilization and ( ii ) phosphate enhances arsenic mobility by stimulating arsenate desorption rather than by inhibiting it at the enzymatic level.
Organizational Affiliation:
Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125.