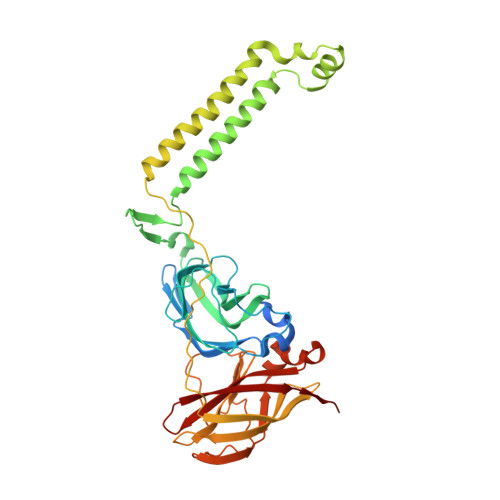
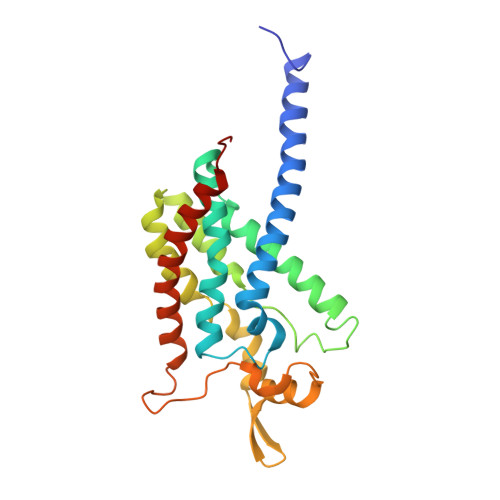
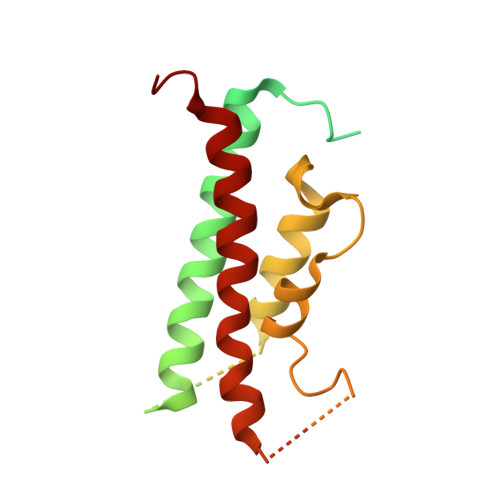
From micelles to bicelles: Effect of the membrane on particulate methane monooxygenase activity.
Ro, S.Y., Ross, M.O., Deng, Y.W., Batelu, S., Lawton, T.J., Hurley, J.D., Stemmler, T.L., Hoffman, B.M., Rosenzweig, A.C.(2018) J Biol Chem 293: 10457-10465
- PubMed: 29739854
- DOI: https://doi.org/10.1074/jbc.RA118.003348
- Primary Citation of Related Structures:
6CXH - PubMed Abstract:
Particulate methane monooxygenase (pMMO) is a copper-dependent integral membrane metalloenzyme that converts methane to methanol in methanotrophic bacteria. Studies of isolated pMMO have been hindered by loss of enzymatic activity upon its removal from the native membrane. To characterize pMMO in a membrane-like environment, we reconstituted pMMOs from Methylococcus ( Mcc. ) capsulatus (Bath) and Methylomicrobium ( Mm. ) alcaliphilum 20Z into bicelles. Reconstitution into bicelles recovers methane oxidation activity lost upon detergent solubilization and purification without substantial alterations to copper content or copper electronic structure, as observed by electron paramagnetic resonance (EPR) spectroscopy. These findings suggest that loss of pMMO activity upon isolation is due to removal from the membranes rather than caused by loss of the catalytic copper ions. A 2.7 Å resolution crystal structure of pMMO from Mm. alcaliphilum 20Z reveals a mononuclear copper center in the PmoB subunit and indicates that the transmembrane PmoC subunit may be conformationally flexible. Finally, results from extended X-ray absorption fine structure (EXAFS) analysis of pMMO from Mm. alcaliphilum 20Z were consistent with the observed monocopper center in the PmoB subunit. These results underscore the importance of studying membrane proteins in a membrane-like environment and provide valuable insight into pMMO function.
Organizational Affiliation:
From the Departments of Molecular Biosciences and Chemistry, Northwestern University, Evanston, Illinois 60208 and.