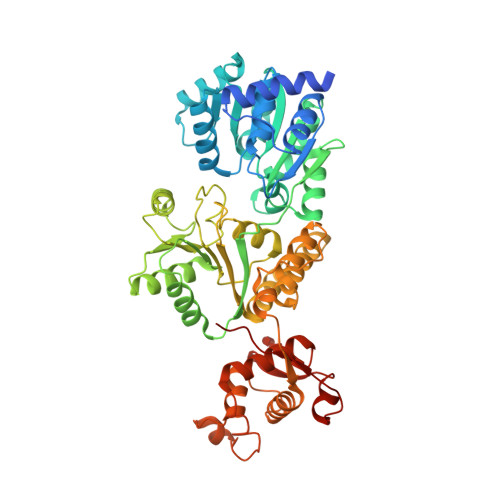
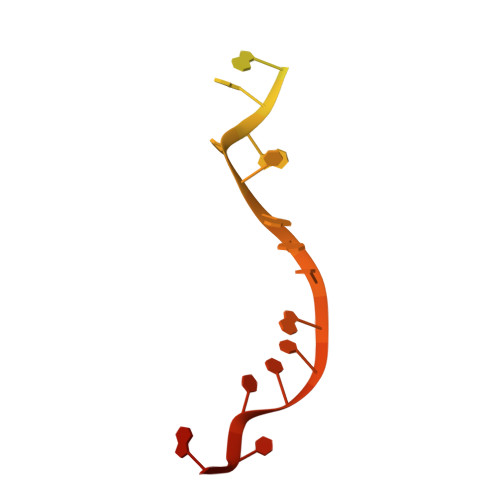
A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases.
Voter, A.F., Qiu, Y., Tippana, R., Myong, S., Keck, J.L.(2018) Nat Commun 9: 4201-4201
- PubMed: 30305632
- DOI: https://doi.org/10.1038/s41467-018-06751-8
- Primary Citation of Related Structures:
6CRM - PubMed Abstract:
Homeostatic regulation of G-quadruplexes (G4s), four-stranded structures that can form in guanine-rich nucleic acids, requires G4 unwinding helicases. The mechanisms that mediate G4 unwinding remain unknown. We report the structure of a bacterial RecQ DNA helicase bound to resolved G4 DNA. Unexpectedly, a guanine base from the unwound G4 is sequestered within a guanine-specific binding pocket. Disruption of the pocket in RecQ blocks G4 unwinding, but not G4 binding or duplex DNA unwinding, indicating its essential role in structure-specific G4 resolution. A novel guanine-flipping and sequestration model that may be applicable to other G4-resolving helicases emerges from these studies.
Organizational Affiliation:
Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, WI, 53706, USA.