Improving the kinetic parameters of nicotine oxidizing enzymes by homologous structure comparison and rational design
Deay, D.O., Seibold, S., Battaile, K.P., Lovell, S., Richter, M.L., Petillo, P.A.(2022) Arch Biochem Biophys : 109122
Experimental Data Snapshot
(2022) Arch Biochem Biophys : 109122
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| (S)-6-hydroxynicotine oxidase | 440 | Shinella sp. HZN7 | Mutation(s): 0 Gene Names: nctB, shn_30305 EC: 1.5.3.5 | 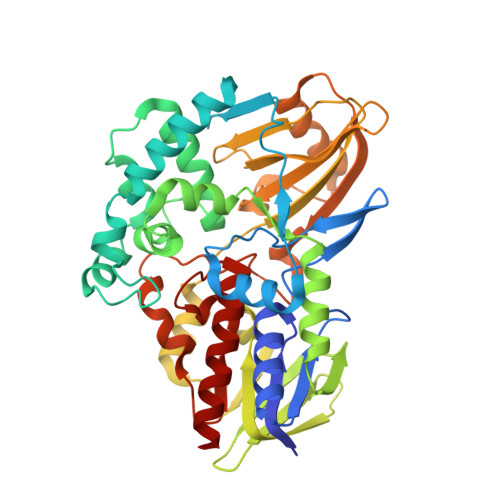 | |
UniProt | |||||
Find proteins for A0A075BSX9 (Shinella sp. (strain HZN7)) Explore A0A075BSX9 Go to UniProtKB: A0A075BSX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A075BSX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | H [auth A] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| MPD Query on MPD | D [auth A], E [auth A], F [auth A] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| ACT Query on ACT | G [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CA Query on CA | B [auth A], C [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.282 | α = 90 |
| b = 96.282 | β = 90 |
| c = 78.628 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| CRANK2 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | P30GM110761 |
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | 2R44DA033701 |