The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B3 protein loaded with linoleic acid to 1.47 Angstrom resolution
Cuypers, M.G., Subramanian, C., White, S.W., Rock, C.O.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fatty Acid Kinase (Fak) B3 protein | A, B [auth D] | 301 | Streptococcus pneumoniae R6 | Mutation(s): 0 Gene Names: spr0652 | 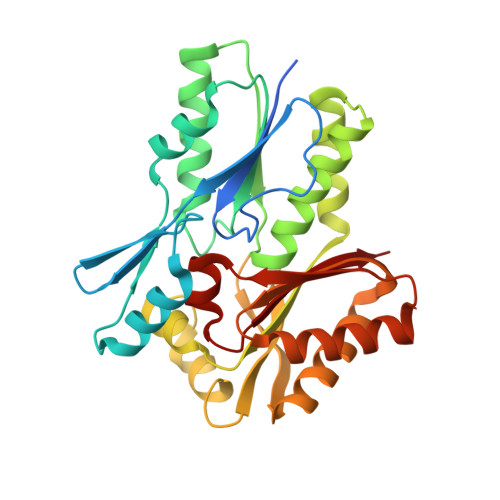 |
UniProt | |||||
Find proteins for Q8DQI6 (Streptococcus pneumoniae (strain ATCC BAA-255 / R6)) Explore Q8DQI6 Go to UniProtKB: Q8DQI6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DQI6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EIC Query on EIC | C [auth A], K [auth D] | LINOLEIC ACID C18 H32 O2 OYHQOLUKZRVURQ-HZJYTTRNSA-N |  | ||
| GOL Query on GOL | E [auth A] F [auth A] G [auth A] H [auth A] M [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | I [auth A], J [auth A], V [auth D], W [auth D], X [auth D] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| NA Query on NA | D [auth A], L [auth D] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 52.775 | α = 90 |
| b = 103.414 | β = 112.99 |
| c = 67.469 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |