The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus.
Mashburn-Warren, L., Goodman, S.D., Federle, M.J., Prehna, G.(2018) Sci Rep 8: 16535-16535
- PubMed: 30409983
- DOI: https://doi.org/10.1038/s41598-018-34816-7
- Primary Citation of Related Structures:
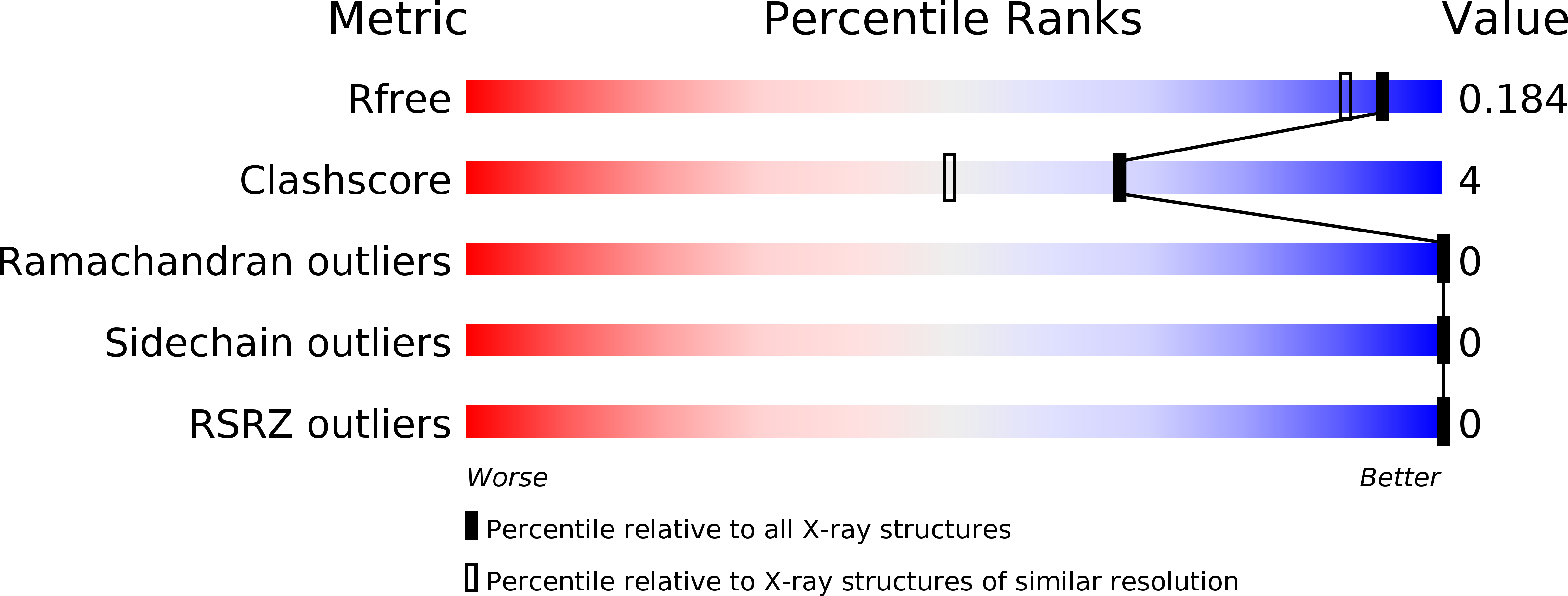
6CKA - PubMed Abstract:
Horizontal gene transfer is an important means of bacterial evolution. This includes natural genetic transformation, where bacterial cells become "competent" and DNA is acquired from the extracellular environment. Natural competence in many species of Streptococcus, is regulated by quorum sensing via the ComRS receptor-signal pair. The ComR-XIP (mature ComS peptide) complex induces expression of the alternative sigma factor SigX, which targets RNA polymerase to CIN-box promoters to activate genes involved in DNA uptake and recombination. In addition, the widely distributed Streptococcus prophage gene paratox (prx) also contains a CIN-box, and here we demonstrate it to be transcriptionally activated by XIP. In vitro experiments demonstrate that Prx binds ComR directly and prevents the ComR-XIP complex from interacting with DNA. Mutations of prx in vivo caused increased expression of the late competence gene ssb when induced with XIP as compared to wild-type, and Prx orthologues are able to inhibit ComR activation by XIP in a reporter strain which lacks an endogenous prx. Additionally, an X-ray crystal structure of Prx reveals a unique fold that implies a novel molecular mechanism to inhibit ComR. Overall, our results suggest Prx functions to inhibit the acquisition of new DNA by Streptococcus.
Organizational Affiliation:
Center for Microbial Pathogenesis, The Research Institute at Nationwide Children's Hospital, Columbus, Ohio, USA.