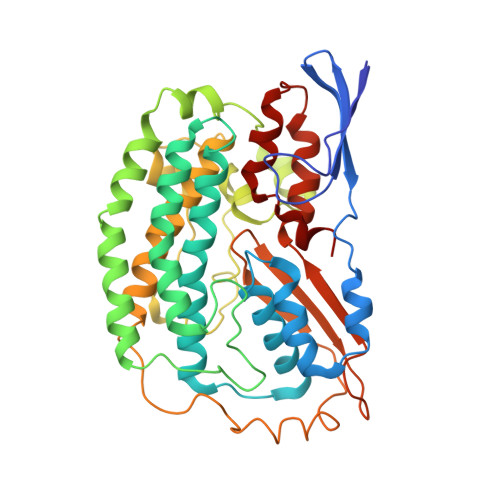
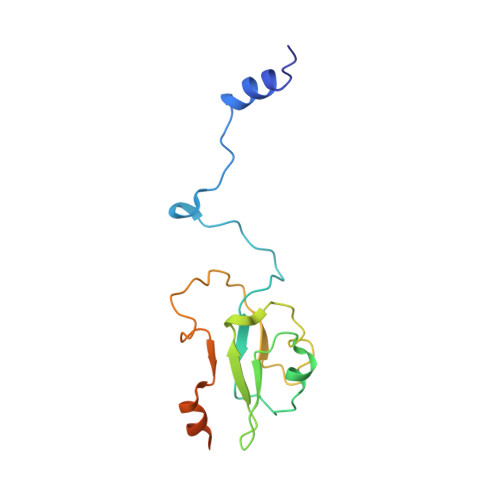
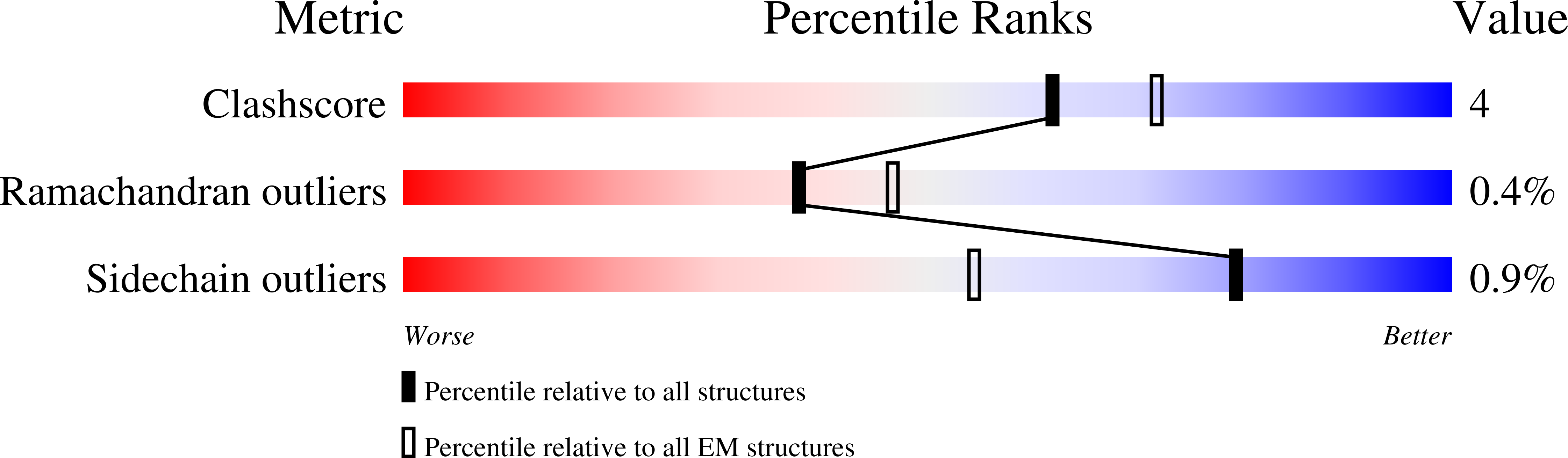Structure of an Ancient Respiratory System.
Yu, H., Wu, C.H., Schut, G.J., Haja, D.K., Zhao, G., Peters, J.W., Adams, M.W.W., Li, H.(2018) Cell 173: 1636
- PubMed: 29754813
- DOI: https://doi.org/10.1016/j.cell.2018.03.071
- Primary Citation of Related Structures:
6CFW - PubMed Abstract:
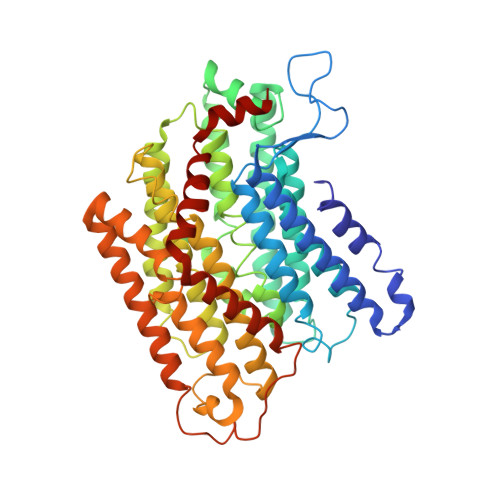
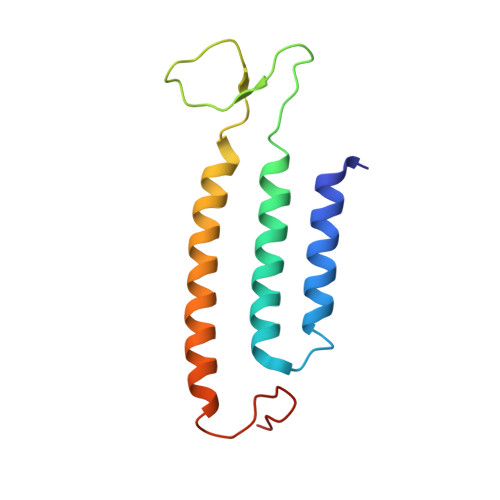
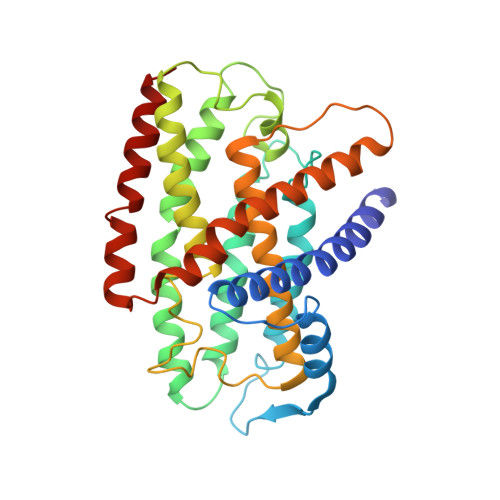
Hydrogen gas-evolving membrane-bound hydrogenase (MBH) and quinone-reducing complex I are homologous respiratory complexes with a common ancestor, but a structural basis for their evolutionary relationship is lacking. Here, we report the cryo-EM structure of a 14-subunit MBH from the hyperthermophile Pyrococcus furiosus. MBH contains a membrane-anchored hydrogenase module that is highly similar structurally to the quinone-binding Q-module of complex I while its membrane-embedded ion-translocation module can be divided into a H + - and a Na + -translocating unit. The H + -translocating unit is rotated 180° in-membrane with respect to its counterpart in complex I, leading to distinctive architectures for the two respiratory systems despite their largely conserved proton-pumping mechanisms. The Na + -translocating unit, absent in complex I, resembles that found in the Mrp H + /Na + antiporter and enables hydrogen gas evolution by MBH to establish a Na + gradient for ATP synthesis near 100°C. MBH also provides insights into Mrp structure and evolution of MBH-based respiratory enzymes.
Organizational Affiliation:
Cryo-EM Structural Biology Laboratory, Van Andel Research Institute, Grand Rapids, MI 49503, USA.