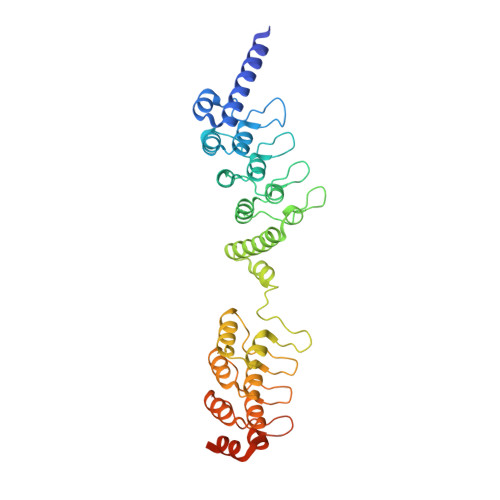
Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
DaRosa, P.A., Klevit, R.E., Xu, W.(2018) Protein Sci 27: 1057-1067
- PubMed: 29604130
- DOI: https://doi.org/10.1002/pro.3413
- Primary Citation of Related Structures:
6CF6 - PubMed Abstract:
Poly(ADP-ribosyl)ation (PARylation) catalyzed by the tankyrase enzymes (Tankyrase-1 and -2; a.k.a. PARP-5a and -5b) is involved in mitosis, telomere length regulation, GLUT-4 vesicle transport, and cell growth and differentiation. Together with the E3 ubiquitin ligase RNF146 (a.k.a. Iduna), tankyrases regulate the cellular levels of several important proteins including Axin, 3BP2, and angiomotins, which are key regulators of Wnt, Src and Hippo signaling, respectively. These tankyrase substrates are first PARylated and then ubiquitylated by RNF146, which is allosterically activated by binding to PAR polymer. Each tankyrase substrate is recognized by a tankyrase-binding motif (TBM). Here we show that RNF146 binds directly to tankyrases via motifs in its C-terminal region. Four of these RNF146 motifs represent novel, extended TBMs, that have one or two additional amino acids between the most conserved Arg and Gly residues. The individual RNF146 motifs display weak binding, but together mediate a strong multivalent interaction with the substrate-binding region of TNKS, forming a robust one-to-one complex. A crystal structure of the first RNF146 noncanonical TBM in complex with the second ankyrin repeat domain of TNKS shows how an extended motif can be accommodated in a peptide-binding groove on tankyrases. Overall, our work demonstrates the existence of a new class of extended TBMs that exist in previously uncharacterized tankyrase-binding proteins including those of IF4A1 and NELFE.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, Washington, 98195.