Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Ruszkowski, M., Sekula, B., Ruszkowska, A., Dauter, Z.(2018) Front Plant Sci 9: 584-584
- PubMed: 29868052
- DOI: https://doi.org/10.3389/fpls.2018.00584
- Primary Citation of Related Structures:
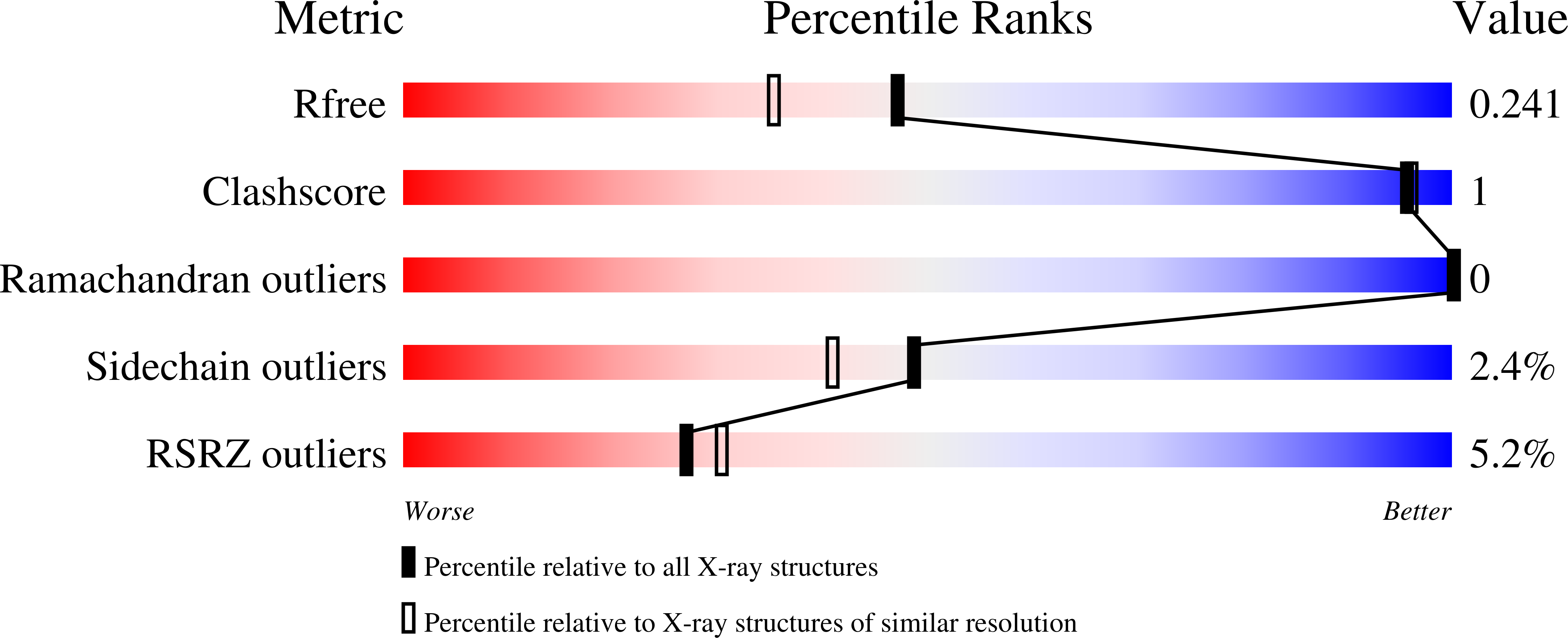
6CCZ, 6CD0, 6CD1 - PubMed Abstract:
Serine hydroxymethyltransferase (SHMT, EC 2.1.2.1) is a pyridoxal 5'-phosphate (PLP)-dependent enzyme which catalyzes the reversible serine-to-glycine conversion in either a tetrahydrofolate-dependent or -independent manner. The enzyme is also responsible for the tetrahydrofolate-independent cleavage of other β-hydroxy amino acids. In addition to being an essential player in the serine homeostasis, SHMT action is the main source of activated one-carbon units, which links SHMT activity with the control of cell proliferation. In plants, studies of SHMT enzymes are more complicated than of those of, e.g., bacterial or mammalian origins because plant genomes encode multiple SHMT isozymes that are targeted to different subcellular compartments: cytosol, mitochondria, plastids, and nucleus. Here we report crystal structures of chloroplast-targeted SHMT from Medicago truncatula ( Mt SHMT3). Mt SHMT3 is a tetramer in solution, composed of two tight and obligate dimers. Our complexes with PLP internal aldimine, PLP-serine and PLP-glycine external aldimines, and PLP internal aldimine with a free glycine reveal structural details of the Mt SHMT3-catalyzed reaction. Capturing the enzyme in different stages along the course of the slow tetrahydrofolate-independent serine-to-glycine conversion allowed to observe a unique conformation of the PLP-serine γ-hydroxyl group, and a concerted movement of two tyrosine residues in the active site.
Organizational Affiliation:
Synchrotron Radiation Research Section of MCL, National Cancer Institute, Argonne, IL, United States.