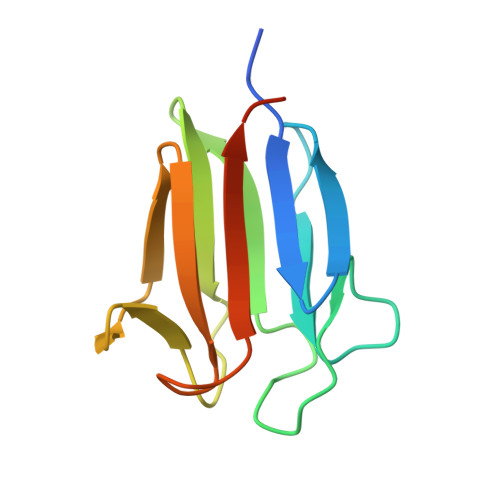
Biophysical Characterization of the Tandem FHA Domain Regulatory Module from the Mycobacterium tuberculosis ABC Transporter Rv1747.
Heinkel, F., Shen, L., Richard-Greenblatt, M., Okon, M., Bui, J.M., Gee, C.L., Gay, L.M., Alber, T., Av-Gay, Y., Gsponer, J., McIntosh, L.P.(2018) Structure 26: 972
- PubMed: 29861345
- DOI: https://doi.org/10.1016/j.str.2018.04.018
- Primary Citation of Related Structures:
6CAH, 6CCD - PubMed Abstract:
The Mycobacterium tuberculosis ATP-binding cassette transporter Rv1747 is a putative exporter of cell wall biosynthesis intermediates. Rv1747 has a cytoplasmic regulatory module consisting of two pThr-interacting Forkhead-associated (FHA) domains connected by a conformationally disordered linker with two phospho-acceptor threonines (pThr). The structures of FHA-1 and FHA-2 were determined by X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, respectively. Relative to the canonical 11-strand β-sandwich FHA domain fold of FHA-1, FHA-2 is circularly permuted and lacking one β-strand. Nevertheless, the two share a conserved pThr-binding cleft. FHA-2 is less stable and more dynamic than FHA-1, yet binds model pThr peptides with moderately higher affinity (∼50 μM versus 500 μM equilibrium dissociation constants). Based on NMR relaxation and chemical shift perturbation measurements, when joined within a polypeptide chain, either FHA domain can bind either linker pThr to form intra- and intermolecular complexes. We hypothesize that this enables tunable phosphorylation-dependent multimerization to regulate Rv1747 transporter activity.
Organizational Affiliation:
Genome Science and Technology Program, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Michael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.