Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
Furlong, E.J., Choudhury, H.G., Kurth, F., Duff, A.P., Whitten, A.E., Martin, J.L.(2018) J Biol Chem 293: 5793-5805
- PubMed: 29491145
- DOI: https://doi.org/10.1074/jbc.RA118.001860
- Primary Citation of Related Structures:
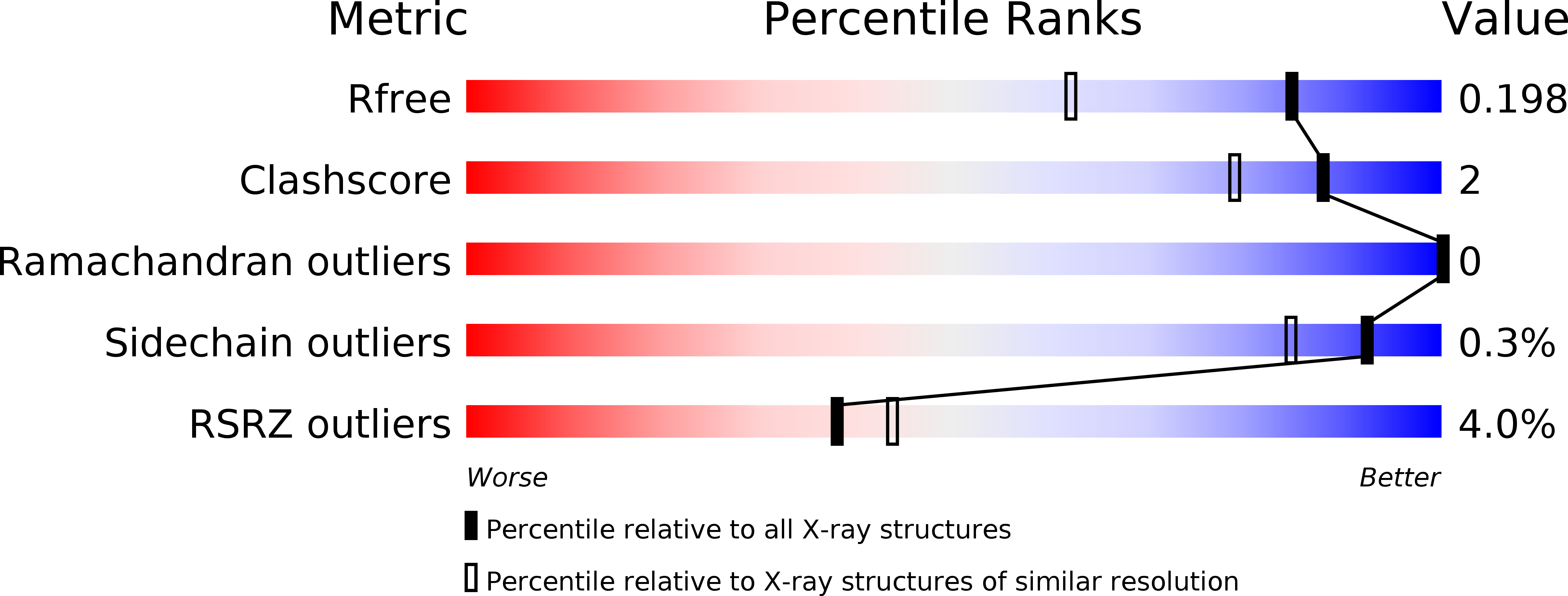
6C29 - PubMed Abstract:
Correct disulfide bond formation is essential for proper folding of many proteins, including bacterial virulence factors. The suppressor of copper sensitivity (Scs) proteins have roles in dithiol/disulfide interchange and the bacterial response to copper stress. Encoded in a four-gene cassette (ScsABCD) present in many Gram-negative bacteria, the Scs proteins are enigmatic and poorly characterized. Here, we show that the periplasmic α-domain of the membrane protein ScsB in the Gram-negative bacterium Proteus mirabilis forms a redox relay with the soluble periplasmic protein PmScsC. We also found that the periplasmic α-domain is sufficient to activate the disulfide isomerase activity of PmScsC. The crystal structure of PmScsBα at a resolution of 1.54 Å revealed that it comprises two structurally similar immunoglobulin-like folds, one of which includes a putative redox-active site with the sequence C XXX C. We confirmed the importance of these cysteine residues for PmScsBα function, and in addition, we engineered cysteine variants that produced a stable complex between PmScsC and PmScsBα. Using small-angle X-ray and neutron scattering analyses with contrast variation, we determined a low-resolution structure of the PmScsC-PmScsBα complex. The structural model of this complex suggested that PmScsBα uses both of its immunoglobulin-like folds to interact with PmScsC and revealed that the highly dynamic PmScsC becomes ordered upon PmScsBα binding. These findings add to our understanding of the poorly characterized Scs proteins.
Organizational Affiliation:
From the Institute for Molecular Bioscience, University of Queensland, St, Lucia, Queensland 4072, Australia.