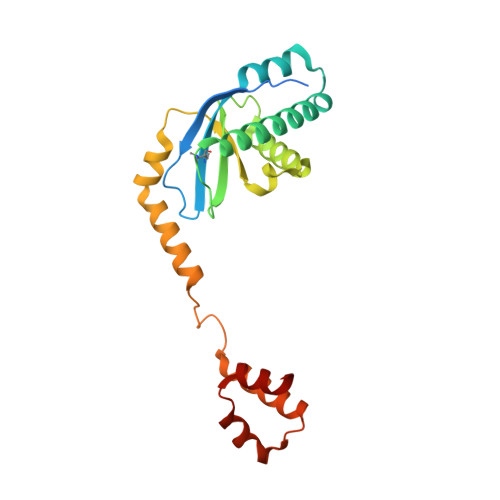
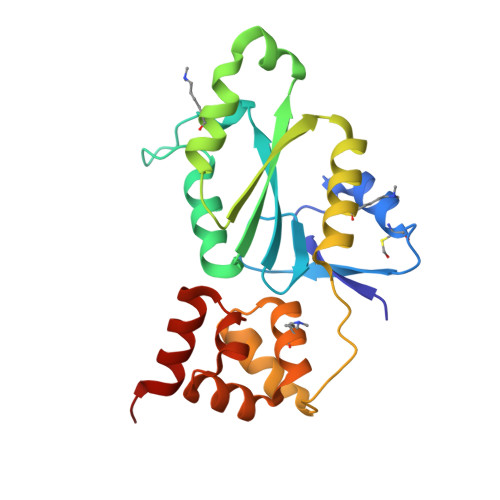
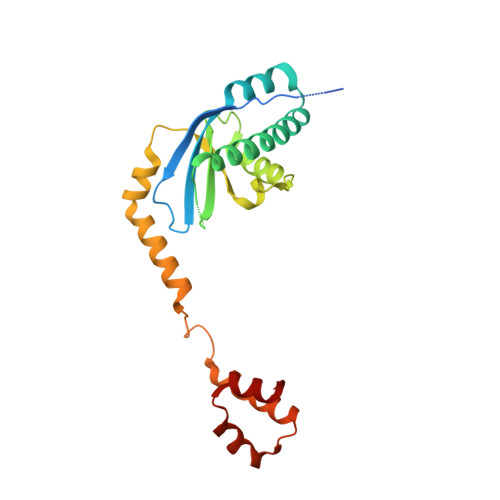
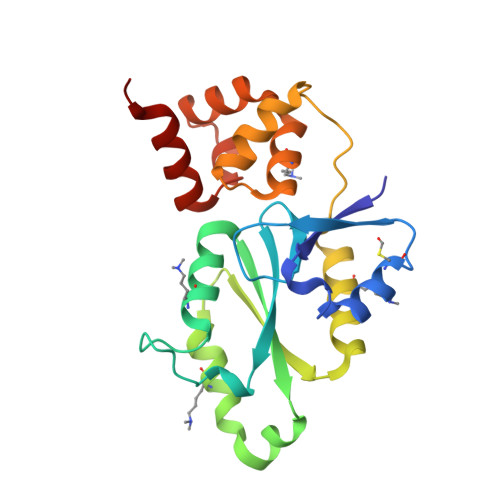
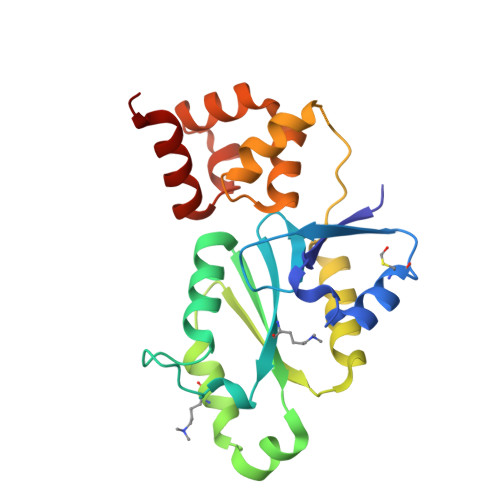
The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Arora, K., Corbett, K.D.(2019) Nucleic Acids Res 47: 2365-2376
- PubMed: 30566683
- DOI: https://doi.org/10.1093/nar/gky1273
- Primary Citation of Related Structures:
6BZF, 6BZG - PubMed Abstract:
In eukaryotic meiosis, generation of haploid gametes depends on the formation of inter-homolog crossovers, which enable the pairing, physical linkage, and eventual segregation of homologs in the meiosis I division. A class of conserved meiosis-specific proteins, collectively termed ZMMs, are required for formation and spatial control of crossovers throughout eukaryotes. Here, we show that three Saccharomyces cerevisiae ZMM proteins-Zip2, Zip4 and Spo16-interact with one another and form a DNA-binding complex critical for crossover formation and control. We determined the crystal structure of a Zip2:Spo16 subcomplex, revealing a heterodimer structurally related to the XPF:ERCC1 endonuclease complex. Zip2:Spo16 lacks an endonuclease active site, but binds specific DNA structures found in early meiotic recombination intermediates. Mutations in multiple DNA-binding surfaces on Zip2:Spo16 severely compromise DNA binding, supporting a model in which the complex's central and HhH domains cooperate to bind DNA. Overall, our data support a model in which the Zip2:Zip4:Spo16 complex binds and stabilizes early meiotic recombination intermediates, then coordinates additional factors to promote crossover formation and license downstream events including synaptonemal complex assembly.
Organizational Affiliation:
Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093, USA.