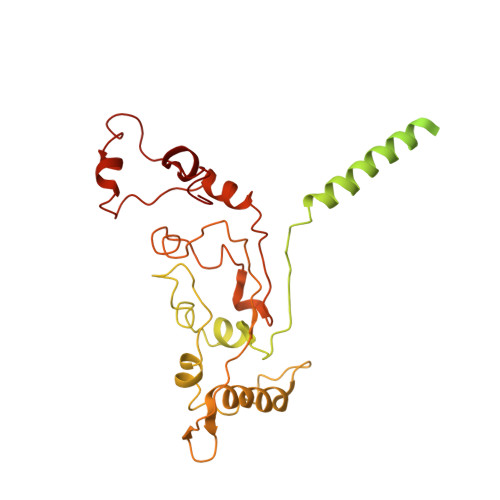
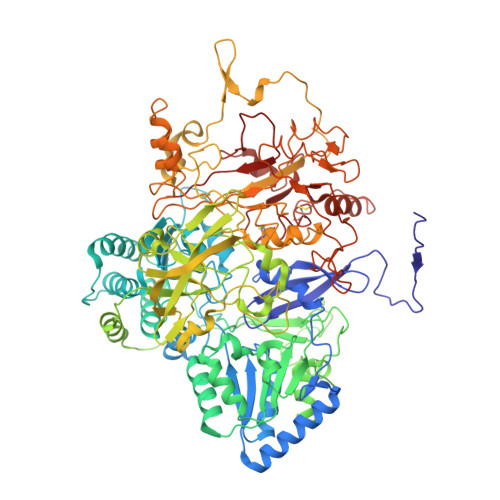
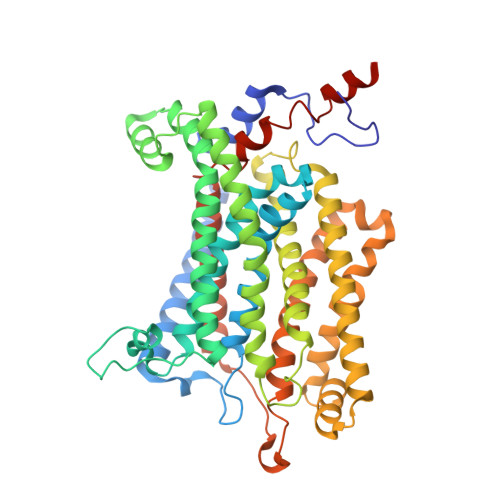
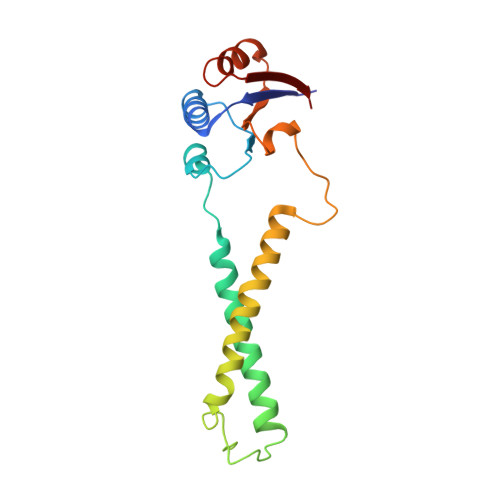
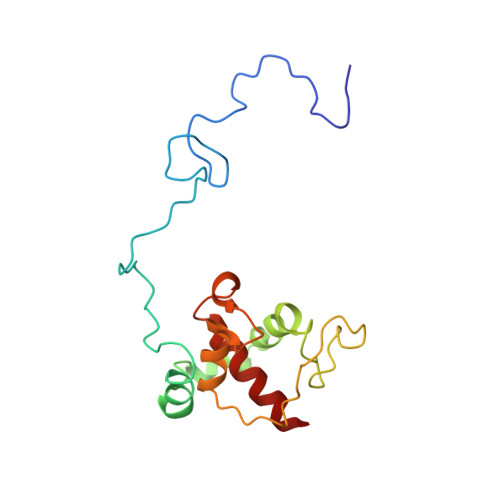
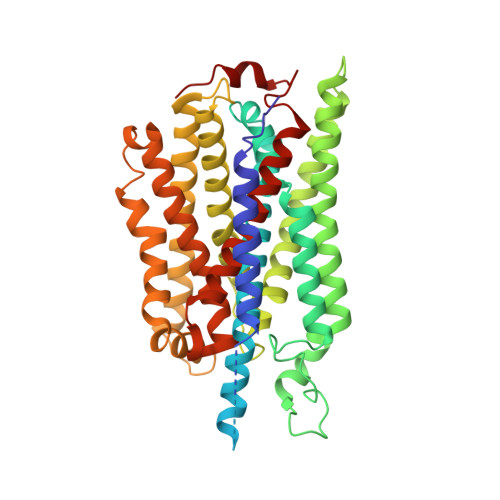
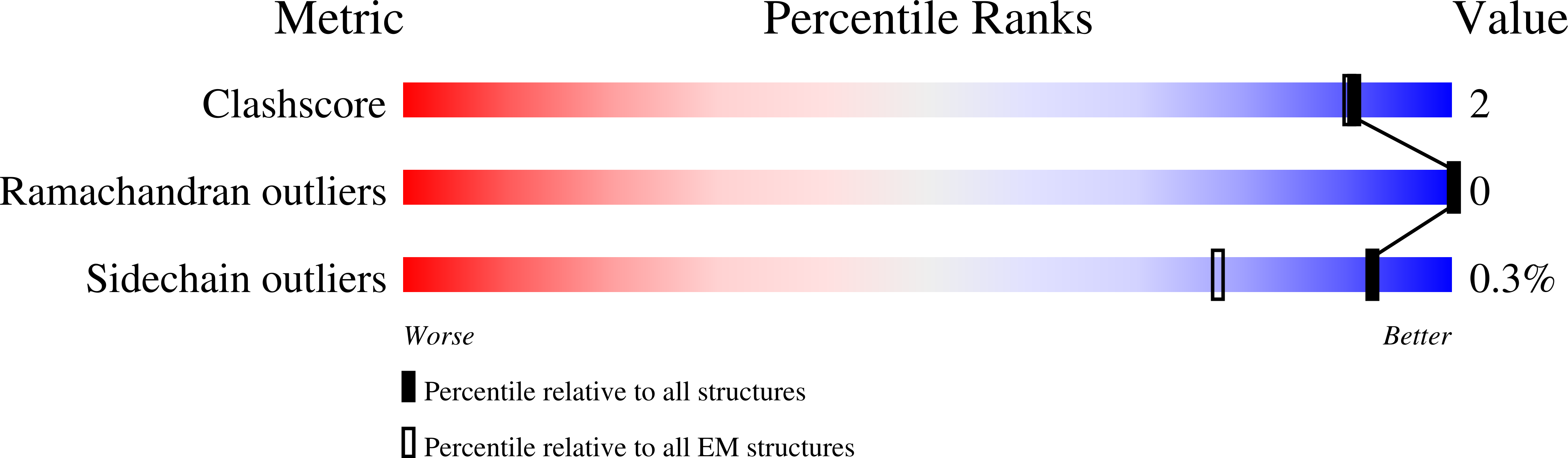Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Sun, C., Benlekbir, S., Venkatakrishnan, P., Wang, Y., Hong, S., Hosler, J., Tajkhorshid, E., Rubinstein, J.L., Gennis, R.B.(2018) Nature 557: 123-126
- PubMed: 29695868
- DOI: https://doi.org/10.1038/s41586-018-0061-y
- Primary Citation of Related Structures:
6BTM - PubMed Abstract:
Alternative complex III (ACIII) is a key component of the respiratory and/or photosynthetic electron transport chains of many bacteria 1-3 . Like complex III (also known as the bc 1 complex), ACIII catalyses the oxidation of membrane-bound quinol and the reduction of cytochrome c or an equivalent electron carrier. However, the two complexes have no structural similarity 4-7 . Although ACIII has eluded structural characterization, several of its subunits are known to be homologous to members of the complex iron-sulfur molybdoenzyme (CISM) superfamily 8 , including the proton pump polysulfide reductase 9,10 . We isolated the ACIII from Flavobacterium johnsoniae with native lipids using styrene maleic acid copolymer 11-14 , both as an independent enzyme and as a functional 1:1 supercomplex with an aa 3 -type cytochrome c oxidase (cyt aa 3 ). We determined the structure of ACIII to 3.4 Å resolution by cryo-electron microscopy and constructed an atomic model for its six subunits. The structure, which contains a [3Fe-4S] cluster, a [4Fe-4S] cluster and six haem c units, shows that ACIII uses known elements from other electron transport complexes arranged in a previously unknown manner. Modelling of the cyt aa 3 component of the supercomplex revealed that it is structurally modified to facilitate association with ACIII, illustrating the importance of the supercomplex in this electron transport chain. The structure also resolves two of the subunits of ACIII that are anchored to the lipid bilayer with N-terminal triacylated cysteine residues, an important post-translational modification found in numerous prokaryotic membrane proteins that has not previously been observed structurally in a lipid bilayer.
Organizational Affiliation:
Department of Biochemistry, University of Illinois, Urbana, IL, USA.