Cork-in-Bottle Occlusion of Fluoride Ion Channels by Crystallization Chaperones.
McIlwain, B.C., Newstead, S., Stockbridge, R.B.(2018) Structure 26: 635-639.e1
- PubMed: 29526432
- DOI: https://doi.org/10.1016/j.str.2018.02.004
- Primary Citation of Related Structures:
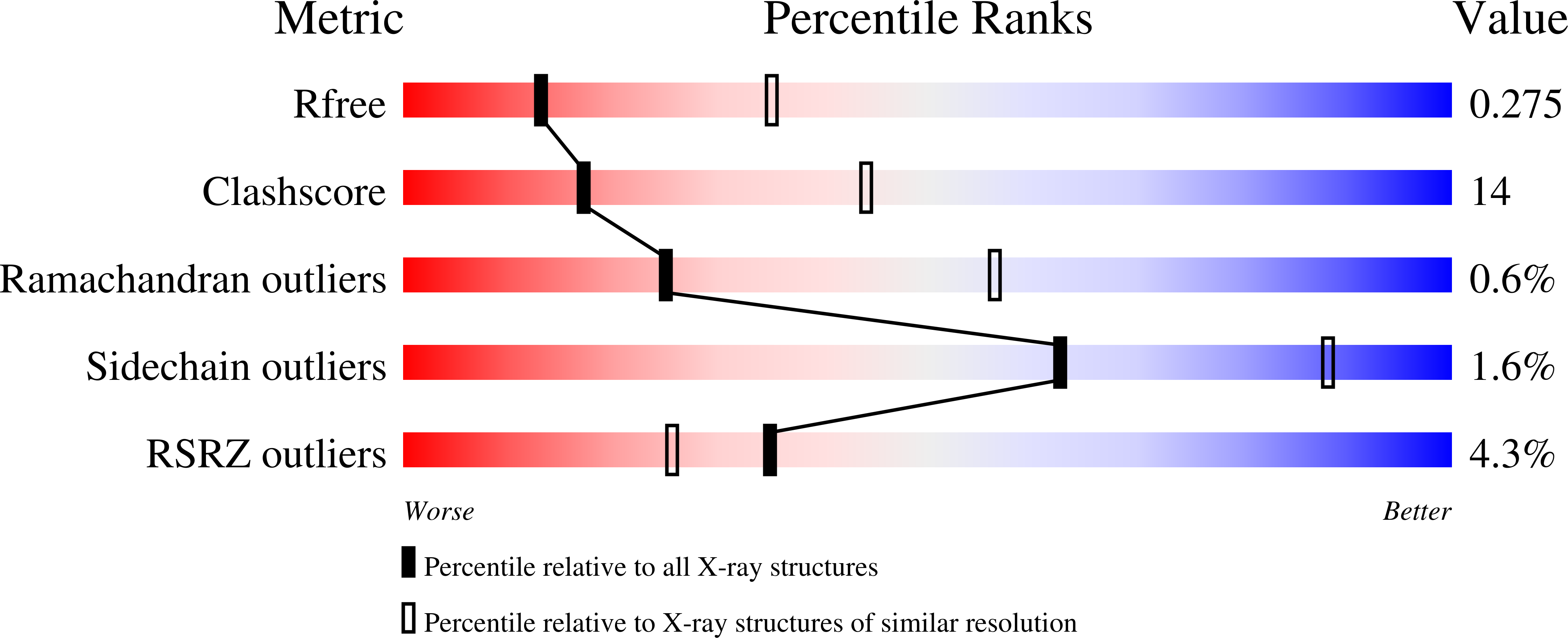
6BQO - PubMed Abstract:
Crystallization of dual-topology fluoride (Fluc) channels requires small, soluble crystallization chaperones known as monobodies, which act as primary crystal lattice contacts. Previous structures of Flucs have been solved in the presence of monobodies that inhibit fluoride currents in single-channel electrophysiological recordings. These structures have revealed two-fold symmetric, doubly bound arrangements, with one monobody on each side of the membrane. The combined electrophysiological and structural observations raise the possibility that chaperone binding allosterically closes the channel, altering the structure from its conducting form. To address this, we identify and solve the structure with a different monobody that only partially blocks fluoride currents. The structure of the channel-monobody complex is asymmetric, with monobody bound to one side of the channel only. The channel conformation is nearly identical on the bound and uncomplexed sides, and to all previously solved structures, providing direct structural evidence that monobody binding does not induce local structural changes.
Organizational Affiliation:
Program in Biophysics, University of Michigan, Ann Arbor, MI 48109, USA.