Stu2 uses a 15-nm parallel coiled coil for kinetochore localization and concomitant regulation of the mitotic spindle.
Haase, K.P., Fox, J.C., Byrnes, A.E., Adikes, R.C., Speed, S.K., Haase, J., Friedman, B., Cook, D.M., Bloom, K., Rusan, N.M., Slep, K.C.(2018) Mol Biol Cell 29: 285-294
- PubMed: 29187574
- DOI: https://doi.org/10.1091/mbc.E17-01-0057
- Primary Citation of Related Structures:
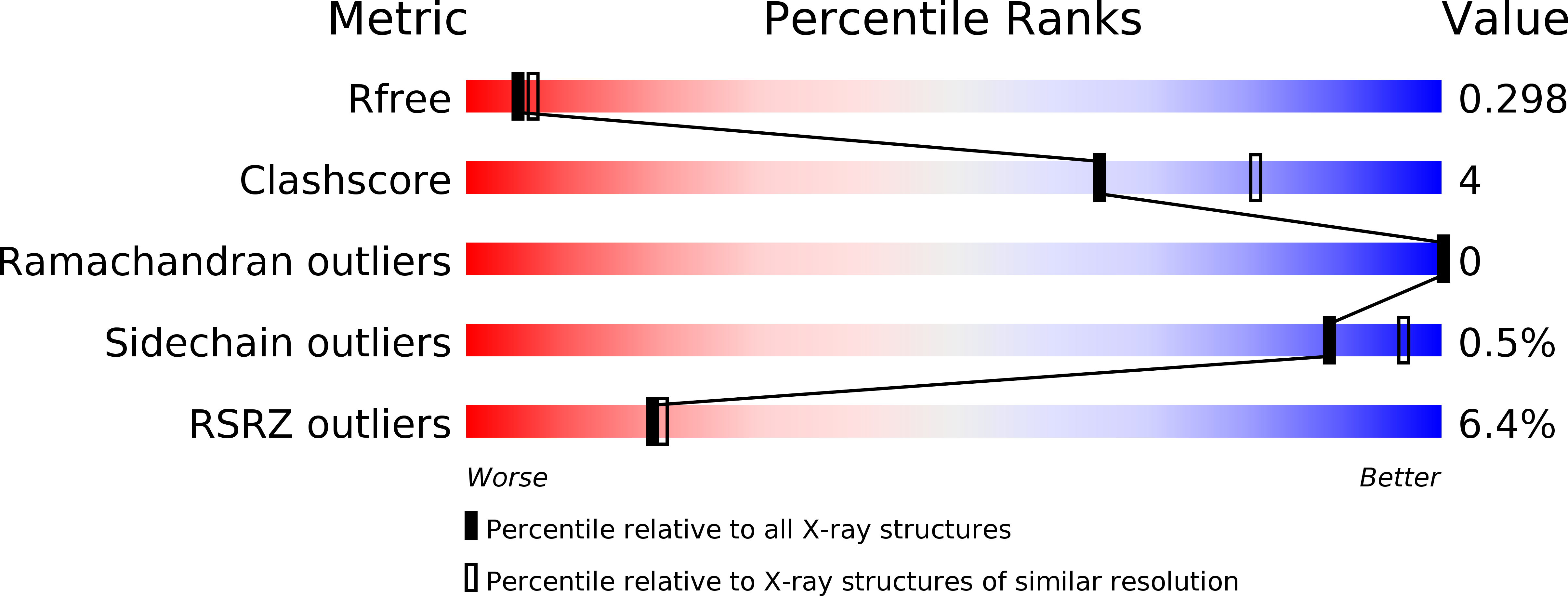
6BL7 - PubMed Abstract:
XMAP215/Dis1 family proteins are potent microtubule polymerases, critical for mitotic spindle structure and dynamics. While microtubule polymerase activity is driven by an N-terminal tumor overexpressed gene (TOG) domain array, proper cellular localization is a requisite for full activity and is mediated by a C-terminal domain. Structural insight into the C-terminal domain's architecture and localization mechanism remain outstanding. We present the crystal structure of the Saccharomyces cerevisiae Stu2 C-terminal domain, revealing a 15-nm parallel homodimeric coiled coil. The parallel architecture of the coiled coil has mechanistic implications for the arrangement of the homodimer's N-terminal TOG domains during microtubule polymerization. The coiled coil has two spatially distinct conserved regions: CRI and CRII. Mutations in CRI and CRII perturb the distribution and localization of Stu2 along the mitotic spindle and yield defects in spindle morphology including increased frequencies of mispositioned and fragmented spindles. Collectively, these data highlight roles for the Stu2 dimerization domain as a scaffold for factor binding that optimally positions Stu2 on the mitotic spindle to promote proper spindle structure and dynamics.
Organizational Affiliation:
Molecular and Cellular Biophysics Program, University of North Carolina, Chapel Hill, NC 27599.