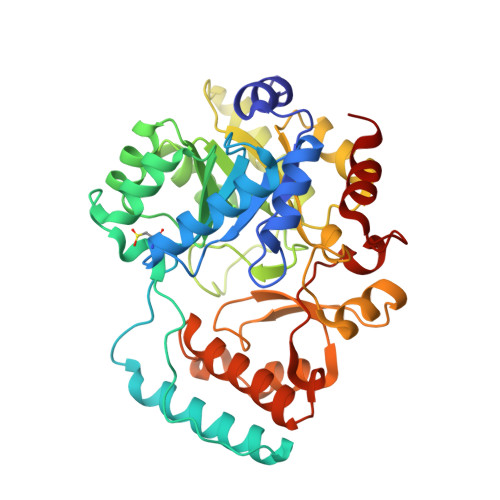
Crystal structure of yeast nitronate monooxygenase from Cyberlindnera saturnus.
Agniswamy, J., Reis, R.A.G., Wang, Y.F., Smitherman, C., Su, D., Weber, I., Gadda, G.(2018) Proteins 86: 599-605
- PubMed: 29383742
- DOI: https://doi.org/10.1002/prot.25470
- Primary Citation of Related Structures:
6BKA - PubMed Abstract:
Nitronate monooxygenase (NMO) is an FMN-dependent enzyme that oxidizes the neurotoxin propionate 3-nitronate (P3N) and represents the best-known system for P3N detoxification in different organisms. The crystal structure of the first eukaryotic Class I NMO from Cyberlindnera saturnus (CsNMO) has been solved at 1.65 Å resolution and refined to an R-factor of 14.0%. The three-dimensional structures of yeast CsNMO and bacterial PaNMO are highly conserved with the exception of three additional loops on the surface in the CsNMO enzyme and differences in four active sites residues. A PEG molecule was identified in the structure and formed extensive interactions with CsNMO, suggesting a specific binding site; however, 8% PEG showed no significant effect on the enzyme activity. This new crystal structure of a eukaryotic NMO provides insight into the function of this class of enzymes.
Organizational Affiliation:
Department of Biology, Georgia State University, Atlanta, Georgia, 30302-3965.