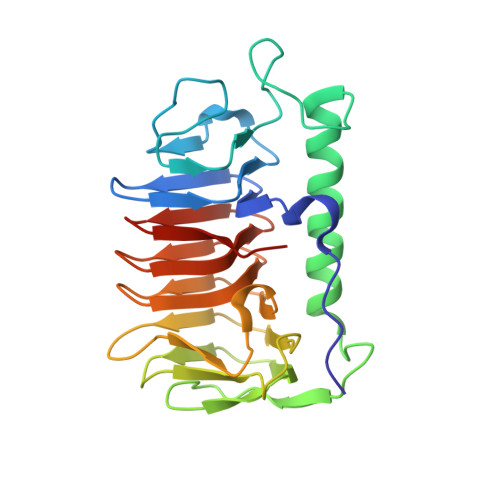
An ice-binding and tandem beta-sandwich domain-containing protein in Shewanella frigidimarina is a potential new type of ice adhesin.
Vance, T.D.R., Graham, L.A., Davies, P.L.(2018) FEBS J 285: 1511-1527
- PubMed: 29498209
- DOI: https://doi.org/10.1111/febs.14424
- Primary Citation of Related Structures:
6BG8 - PubMed Abstract:
Out of the dozen different ice-binding protein (IBP) structures known, the DUF3494 domain is the most widespread, having been passed many times between prokaryotic and eukaryotic microorganisms by horizontal gene transfer. This ~25-kDa β-solenoid domain with an adjacent parallel α-helix is most commonly associated with an N-terminal secretory signal peptide. However, examples of the DUF3494 domain preceded by tandem Bacterial Immunoglobulin-like (BIg) domains are sometimes found, though uncharacterized. Here, we present one such protein (SfIBP_1) from the Antarctic bacterium Shewanella frigidimarina. We have confirmed and characterized the ice-binding activity of its ice-binding domain using thermal hysteresis measurements, fluorescent ice plane affinity analysis, and ice recrystallization inhibition assays. X-ray crystallography was used to solve the structure of the SfIBP_1 ice-binding domain, to further characterize its ice-binding surface and unique method of stabilizing or 'capping' the ends of the solenoid structure. The latter is formed from the interaction of two loops mediated by a combination of tandem prolines and electrostatic interactions. Furthermore, given their domain architecture and membrane association, we propose that these BIg-containing DUF3494 IBPs serve as ice-binding adhesion proteins that are capable of adsorbing their host bacterium onto ice.
Organizational Affiliation:
Department of Biomedical and Molecular Science, Queen's University, Kingston, Canada.