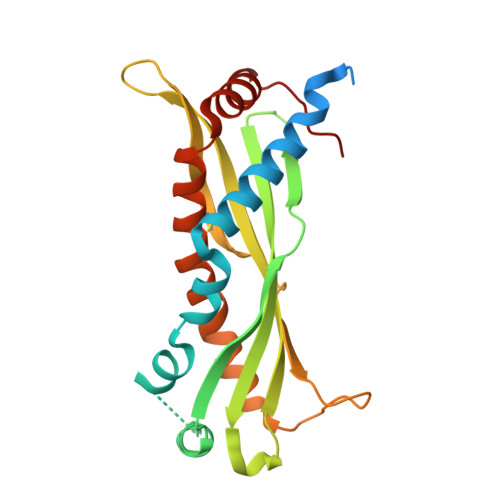
Crystal structure of the mouse innate immunity factor bacterial permeability-increasing family member A1.
Little, M.S., Redinbo, M.R.(2018) Acta Crystallogr F Struct Biol Commun 74: 268-276
- PubMed: 29717993
- DOI: https://doi.org/10.1107/S2053230X18004600
- Primary Citation of Related Structures:
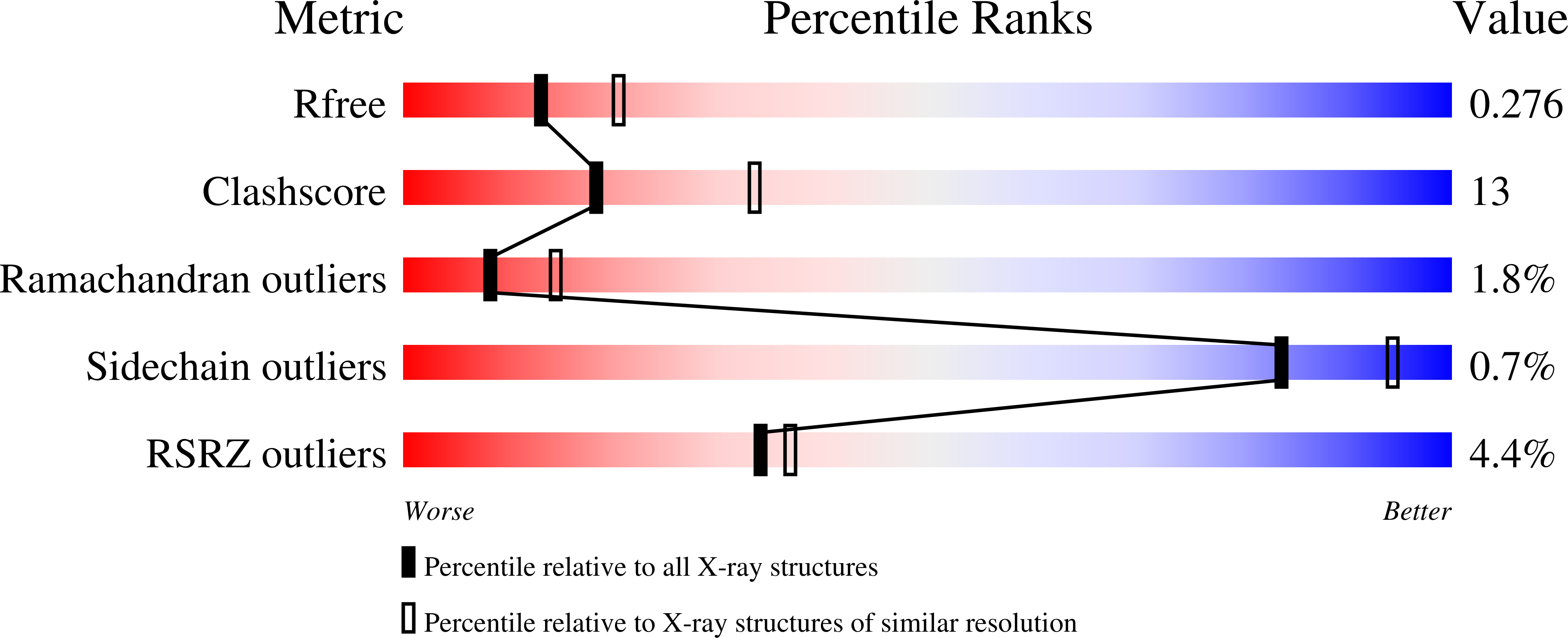
6BAQ - PubMed Abstract:
Bacterial permeability-increasing family member A1 (BPIFA1) is an innate immunity factor and one of the most abundantly secreted proteins in the upper airways. BPIFA1 is multifunctional, with antimicrobial, surfactant and lipopolysaccharide-binding activities, as well as established roles in lung hydration. Here, the 2.5 Å resolution crystal structure of BPIFA1 from Mus musculus (mBPIFA1) is presented and compared with those of human BPIFA1 (hBPIFA1) and structural homologs. Structural distinctions between mBPIFA1 and hBPIFA1 suggest potential differences in biological function, including the regulation of a key pulmonary ion channel.
Organizational Affiliation:
Department of Chemistry, University of North Carolina, 4350 Genome Sciences Building, Chapel Hill, NC 27599-3290, USA.